scRNA-Seq Analysis
Hands-On Excercises
Åsa Björklund
15-Jun-2018
This is the hands-on material for Single-cell RNA sequencing analyses. Below are links to some tutorials for working with scRNAseq data analysis.
1 Introduction
There are multiple packages dedicated to scRNAseq analysis. If you want to try out more exercises there are more tutorials at our 3-day course at:
https://nbisweden.github.io/workshop-scRNAseq/
In this workshop, you have the chance to test two commonly used pipelines:
Seurat package which has implemented all steps of scRNAseq analysis, from QC to clustering and differential expression.
Scater package + SC3. Scater defines the
SingleCellExperimentclass which is a good package for QC. Building on the Scater SCE class, SC3 can be used for clustering.
We suggest that you start with one of these pipelines and see how far you get.
2 The dataset
In all of the exercises, you will be working on a dataset with Smartseq2 data from human tonsil innate lymphoid cells (ILCs) from Björklund et. al., 2016. Since this dataset has been index-sorted with FACS, we already have an idea about what the main celltypes are supposed to be, so it is a good example dataset for evaluating the clustering of our data.
All data can be accessed here.
The ILCs can be subdivided into 3 mainsubpopulations; ILC1, ILC2 and ILC3. Also natural killer cells (NK-cells) were included in the sorting. Samples were taken from 3 individual donors and sorted and prepared in 3 different batches on multiple plates.
The initial plates were gated using known surface markers for the ILCs and NK cells. As the ICL3 is the most abundant population among the ILCs, these plates mainly consists of ILC3s. To increase the representation of the other two subtypes some plates were sorted with specific gating for ILC1s and ILC2s.
Below is a representation of the abundance of the 4 different celltypes on each plate.
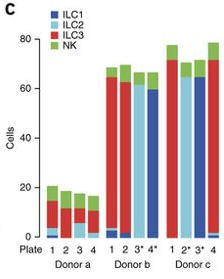
In this dataset there is clearly a batch effect with regards to the different donors and sort occasions. There is also a clear plate effect. But since the cell types differ on the plates, there is not much we can do to distinguish the plate batch effect from the biological variation.
3 Labs
3.1 Seurat
Single-cell RNA-seq data workflow with Seurat including filtering, QC and clustering.
Click here.
3.3 SC3
Single-cell RNA-seq concensus clustering and with the SC3 package.
OBS! It is recommended that you run the Scater lab first that takes you through the step of batch correction before running SC3.
Click here.
4 DLL issues
Many of the scRNAseq packages have a heap of different dependencies. There is a limit to how many libraries you can load at the same time, and when loading a new library you may run into an error message like:
Error: package or namespace load failed for Seurat in dyn.load(file, DLLpath = DLLpath, ...):
unable to load shared object '/Library/Frameworks/R.framework/Versions/3.4/Resources/library/tclust/libs/tclust.so':
` maximal number of DLLs reached...
Error: package or namespace load failed for Seurat in dyn.load(file, DLLpath = DLLpath, ...):If this happens, the easiest solution is to start a new R session and start again from scratch. Another solution is to unload some of the unnecessary loaded packages. You can also set the maximal number of DLLs with the environment variable R_MAX_NUM_DLLS to a large value like R_MAX_NUM_DLLS=500 and restart R.
On a Mac, modify the file /Library/Frameworks/R.framework/Resources/etc/Renviron and add R_MAX_NUM_DLLS=500 in the end. For other OS check https://stat.ethz.ch/R-manual/R-patched/library/base/html/Startup.html.
5 Session Info
- This document has been created in RStudio using R Markdown and related packages.
- For R Markdown, see http://rmarkdown.rstudio.com
- For details about the OS, packages and versions, see detailed information below:
sessionInfo()## R version 3.4.3 (2017-11-30)
## Platform: x86_64-w64-mingw32/x64 (64-bit)
## Running under: Windows >= 8 x64 (build 9200)
##
## Matrix products: default
##
## locale:
## [1] LC_COLLATE=English_United Kingdom.1252
## [2] LC_CTYPE=English_United Kingdom.1252
## [3] LC_MONETARY=English_United Kingdom.1252
## [4] LC_NUMERIC=C
## [5] LC_TIME=English_United Kingdom.1252
##
## attached base packages:
## [1] stats4 parallel stats graphics grDevices utils datasets
## [8] methods base
##
## other attached packages:
## [1] SC3_1.7.7 sva_3.26.0
## [3] BiocParallel_1.12.0 genefilter_1.60.0
## [5] mgcv_1.8-23 nlme_3.1-137
## [7] scater_1.6.3 SingleCellExperiment_1.0.0
## [9] SummarizedExperiment_1.8.1 DelayedArray_0.4.1
## [11] matrixStats_0.53.1 GenomicRanges_1.30.3
## [13] GenomeInfoDb_1.14.0 IRanges_2.12.0
## [15] S4Vectors_0.16.0 Biobase_2.38.0
## [17] BiocGenerics_0.24.0 bindrcpp_0.2.2
## [19] gridExtra_2.3 Seurat_2.3.1
## [21] Matrix_1.2-14 cowplot_0.9.2
## [23] forcats_0.3.0 stringr_1.3.1
## [25] dplyr_0.7.5 purrr_0.2.5
## [27] readr_1.1.1 tidyr_0.8.1
## [29] tibble_1.4.2 ggplot2_2.2.1
## [31] tidyverse_1.2.1 captioner_2.2.3
## [33] bookdown_0.7 knitr_1.20
##
## loaded via a namespace (and not attached):
## [1] DRR_0.0.3 vcd_1.4-4 Hmisc_4.1-1
## [4] ica_1.0-2 RcppRoll_0.3.0 class_7.3-14
## [7] foreach_1.4.4 lmtest_0.9-36 rprojroot_1.3-2
## [10] crayon_1.3.4 laeken_0.4.6 MASS_7.3-50
## [13] WriteXLS_4.0.0 magic_1.5-8 backports_1.1.2
## [16] rlang_0.2.1 XVector_0.18.0 caret_6.0-80
## [19] ROCR_1.0-7 readxl_1.1.0 irlba_2.3.2
## [22] SparseM_1.77 nloptr_1.0.4 limma_3.34.9
## [25] smoother_1.1 rjson_0.2.20 bit64_0.9-7
## [28] glue_1.2.0 pheatmap_1.0.10 trimcluster_0.1-2
## [31] rngtools_1.3.1 sfsmisc_1.1-2 vipor_0.4.5
## [34] AnnotationDbi_1.40.0 shinydashboard_0.7.0 VGAM_1.0-5
## [37] haven_1.1.1 tidyselect_0.2.4 NADA_1.6-1
## [40] rio_0.5.10 fitdistrplus_1.0-9 XML_3.98-1.11
## [43] zoo_1.8-2 xtable_1.8-2 lars_1.2
## [46] MatrixModels_0.4-1 magrittr_1.5 evaluate_0.10.1
## [49] tclust_1.4-1 bibtex_0.4.2 cli_1.0.0
## [52] zlibbioc_1.24.0 rstudioapi_0.7 doRNG_1.6.6
## [55] sp_1.3-1 robCompositions_2.0.8 rpart_4.1-13
## [58] pls_2.6-0 zCompositions_1.1.1 RcppEigen_0.3.3.4.0
## [61] shiny_1.0.5 xfun_0.1 cluster_2.0.7-1
## [64] caTools_1.17.1 sgeostat_1.0-27 doSNOW_1.0.16
## [67] ape_5.1 png_0.1-7 reshape_0.8.7
## [70] ipred_0.9-6 withr_2.1.2 bitops_1.0-6
## [73] ranger_0.10.1 plyr_1.8.4 cellranger_1.1.0
## [76] pcaPP_1.9-73 e1071_1.6-8 pillar_1.2.3
## [79] gplots_3.0.1 mvoutlier_2.0.9 flexmix_2.3-14
## [82] kernlab_0.9-26 scatterplot3d_0.3-41 TTR_0.23-3
## [85] xts_0.10-2 lava_1.6.1 metap_0.9
## [88] tools_3.4.3 ddalpha_1.3.3 foreign_0.8-70
## [91] beeswarm_0.2.3 munsell_0.5.0 proxy_0.4-22
## [94] compiler_3.4.3 abind_1.4-5 httpuv_1.4.3
## [97] dimRed_0.1.0 segmented_0.5-3.0 pkgmaker_0.27
## [100] GenomeInfoDbData_1.0.0 prodlim_2018.04.18 edgeR_3.20.9
## [103] lattice_0.20-35 snow_0.4-2 later_0.7.3
## [106] recipes_0.1.2 jsonlite_1.5 GGally_1.4.0
## [109] scales_0.5.0.9000 pbapply_1.3-4 carData_3.0-1
## [112] lazyeval_0.2.1 promises_1.0.1 car_3.0-0
## [115] doParallel_1.0.11 latticeExtra_0.6-28 R.utils_2.6.0
## [118] reticulate_1.8 checkmate_1.8.5 rmarkdown_1.10
## [121] openxlsx_4.1.0 CVST_0.2-2 Rtsne_0.13
## [124] igraph_1.2.1 survival_2.42-3 yaml_2.1.19
## [127] prabclus_2.2-6 htmltools_0.3.6 memoise_1.1.0
## [130] modeltools_0.2-21 locfit_1.5-9.1 destiny_2.6.2
## [133] viridisLite_0.3.0 digest_0.6.15 rrcov_1.4-4
## [136] assertthat_0.2.0 mime_0.5 registry_0.5
## [139] RSQLite_2.1.1 data.table_1.11.4 blob_1.1.1
## [142] geometry_0.3-6 R.oo_1.22.0 splines_3.4.3
## [145] Formula_1.2-3 labeling_0.3 fpc_2.1-11
## [148] Cairo_1.5-9 mixtools_1.1.0 RCurl_1.95-4.10
## [151] cvTools_0.3.2 broom_0.4.4 hms_0.4.2
## [154] modelr_0.1.2 rhdf5_2.22.0 colorspace_1.3-2
## [157] base64enc_0.1-3 mnormt_1.5-5 ggbeeswarm_0.6.0
## [160] SDMTools_1.1-221 nnet_7.3-12 diffusionMap_1.1-0
## [163] tximport_1.6.0 Rcpp_0.12.17 mclust_5.4
## [166] RANN_2.5.1 mvtnorm_1.0-8 VIM_4.7.0
## [169] truncnorm_1.0-8 ModelMetrics_1.1.0 R6_2.2.2
## [172] grid_3.4.3 ggridges_0.5.0 acepack_1.4.1
## [175] zip_1.0.0 curl_3.2 minqa_1.2.4
## [178] gdata_2.18.0 robustbase_0.93-0 RColorBrewer_1.1-2
## [181] iterators_1.0.9 gower_0.1.2 htmlwidgets_1.2
## [184] biomaRt_2.34.2 rvest_0.3.2 htmlTable_1.12
## [187] codetools_0.2-15 dtw_1.20-1 lubridate_1.7.4
## [190] FNN_1.1 gtools_3.5.0 prettyunits_1.0.2
## [193] psych_1.8.4 R.methodsS3_1.7.1 gtable_0.2.0
## [196] tsne_0.1-3 DBI_1.0.0 httr_1.3.1
## [199] KernSmooth_2.23-15 stringi_1.2.3 progress_1.1.2
## [202] reshape2_1.4.3 diptest_0.75-7 annotate_1.56.2
## [205] viridis_0.5.1 timeDate_3043.102 xml2_1.2.0
## [208] boot_1.3-20 lme4_1.1-17 sROC_0.1-2
## [211] DEoptimR_1.0-8 bit_1.1-14 pkgconfig_2.0.1
## [214] bindr_0.1.1Page built on: 15-Jun-2018 at 22:19:54.
2018 | SciLifeLab > NBIS > RaukR