 Tips for Spatial Transcriptomics projects
Tips for Spatial Transcriptomics projects
Here, we will try to list some issues, tips related to different programs etc.
Loading data
Loading h5ad in R
To convert a scanpy AnnData object to a Seurat object in R, you need to have SeuratDisk installed. It should then be easy to read it in in R, however, it is very sensitive to having the correct formats, naming conventions etc for it to work. So it may at times require some troubleshooting.
Loading the VISp scRNAseq dataset should work with:
library(Seurat)
library(SeuratDisk)
library(hdf5r)
Convert("VISp-sc_data.h5ad", dest = "h5seurat", overwrite = TRUE)
data <- LoadH5Seurat("VISp-sc_data.h5seurat")
Scanpy hints
Loading data
A good idea is to specify library_id when you load visium data so that you have easy to interpret names of your images that matches library_id in the metadata (adata.obs):
adata = sc.read_visium("../data/mouse_brain_visium_wo_cloupe_data/rawdata/ST8059049/", library_id = "ST8059049")
Barplots
Matplotlib (https://matplotlib.org/) has some conventient functions for plotting, for instance you can plot sample distribution per cluster with:
tmp = pd.crosstab(adata.obs['clusters'],adata.obs['library_id'], normalize='index')
tmp.plot.bar(stacked=True).legend(loc='upper right')
Selection of image region with Napari
Some example code on how to select part of of a section in folder 'notebooks/napari_test.ipynb'
Reverting back to raw matrix
If you want the full lognormalized (or whatever you stored in adata.raw) matrix again:
adata = adata.raw.to_adata()
Convert from dense to sparse and back
Some packages may require the matrices to be in a specific format, and hence will not run if you have a dense/sparse matrix, but it is easy to convert them.
Sparse to dense:
adata.X.todense()
Dense to sparse (you can use either csr or rsc (column to row or row to column)
from scipy.sparse import csr_matrix
adata.X = csr_matrix(adata.X)
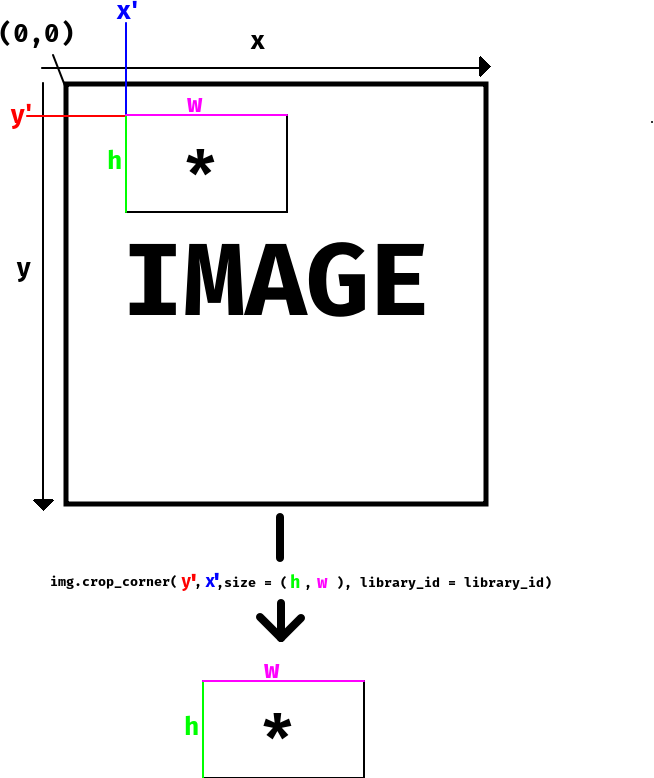
Cropping data with squidpy
If we have an image contained within an ImageContainer object called img in
squidpy, we can crop this image by using the function crop_corner (there are
other methods as well). You can read the documentation for this function
here,
but in short you use the following command to crop your image:
crop = img.crop_corner(y,x,size = (h,w),library_id = library_id)
which will give you a new ImageContainer object called crop. Below we've also
assembled an image that explains how the selection works and how the different
parameters (y,x,h and w) relates to the original and cropped image.
Running tangram, nuclei segmentation and image features:
- Some example code on how to run tangram in 'notebooks/tangram_test.ipynb'
- Some example code on how to run nuceli segmentation and then tangram, and also calculate image features, in 'notebooks/tangram_test_segmentation.ipynb'
Selection of image region with Napari
Some example code on how to select part of of a section in folder 'notebooks/napari_test.ipynb'