 Pre-course Materials
Pre-course Materials
This workshop is aimed towards biologists, researchers, computer scientists or data analysts planning to run, analyse and interpret single cells RNA-seq experiments independently. Basic knowledge of working on the Unix/Linux command line, R/Python and RNA-seq is therefore expected.
Please follow the instructions below for installations prior to the workshop:
- Knowledge Requirements
- Slack
- Zoom
- AWS instance
- Conda
- Dataset
- Code
 Knowledge Requirements
Knowledge Requirements
We strongly recommend for those not yet familiar with UNIX and/or R/Python to take this opportunity and take these online tutorials, since those are requirements for the workshop. This will help you to develop your programming skills and we can always learn a few tricks here and there, even if you are already experienced.
- UNIX (part_1): http://swcarpentry.github.io/shell-novice/
- UNIX (part_2): https://carpentries-incubator.github.io/shell-extras/
- R (part_1): https://swcarpentry.github.io/r-novice-inflammation/
- R (part_2): http://swcarpentry.github.io/r-novice-gapminder/
- Python (part_1):https://swcarpentry.github.io/python-novice-inflammation/
- Python (part_2): http://swcarpentry.github.io/python-novice-gapminder/
After taking those courses (or any other equivalent course in programming in bash and R or Python) will provide you with the basics in, for example:
- file structure and manipulation in bash
- loading, handling and manipulating vectors, matrices, factors and lists
- creating for-loops
- using Rmarkdown/Jupyter for reports
- editing and writing files in the command line
- and much more ...
 Slack
Slack
Make sure that you have Slack installed because we will use it a lot during the workshop. Communication, troubleshooting and group discussions will happen via Slack workspace single-cellschool2021. All accepted students will receive an invitation link via email. Please add this workspace to your Slack application on your desktop and do NOT use it in the web.
Please join the following channels once you are in the workspace:
#software-to-installfor questions about installation and troubleshooting#generalfor general questions about the workshop
Note: Please post your question in the channel and NOT directly to the teacher. Any participant that knows the answer to any problem is encouraged to help too.
 Zoom
Zoom
Make sure that you have the latest Zoom (version 5.7.0 or above) installed because we will use it a lot during the workshop for the lecture and groups discussions.
Previous Zoom versions will not work. If you already have Zoom installed, you can also update it by clicking in Check for Updates ... button in the main top menu.
 AWS instance
AWS instance
You will be provided access to a cloud instance containing all necessary installations and computational infrastructure at the start of the course. To work on the cloud instance the only program you need will be a modern browser (i.e. Firefox, Edge, Chrome, Safari). You will connect either through Rstudio server or Jupyter lab. If you would like to work on your own computer, read the instructions on installing dependencies with conda locally below (due to specific requirements the instructions below will not be sufficient for the projects on multi-omics and deep learning).
Login
You will be provided individual login details at the start of the course.
Setup
You will have access to three important directories, that can be approached with the terminal (both Rstudio and jupyter notebook contain an interface to run commands on the terminal):
- Your home directory (at
~): only visible for you and readable and writeable - A group work directory (at
/group_work): visible for everybody, and readable and writable for everybody. If you are using this for your group work, make a subdirectory that belong to your group - A data directory (at
/data): visible for everybody and only readable. This directory contains data that is used for the exercises.
All directories are available from both the jupyter notebook and rstudio interfaces.
You are limited to using 4 CPU and 16 GB of RAM. There is no swap, so if you exceed that limit your environment will crash and restart automatically. In this case, data in memory might be lost, data stored on disk will remain.
We will close down the cloud instance during the night. Everything that is in the working memory will be lost. Therefore, make sure that by the end of the day you store everything that you want to keep on disk.
Rstudio server
The Rstudio server contains an interface for most r-based analyses (e.g. based on R markdown (.Rmd) files). You can install additional packages from CRAN or bioconductor like you are used to do. There is also a possibility to use python packages with reticulate. The environment managing software conda is not available for rstudio server, so if you are installing python packages use a virtualenv. You can e.g. install scanorama like this:
library(reticulate)
system('pipenv --python 3.8')
venv <- system("pipenv --venv", inter = TRUE)
reticulate::use_virtualenv(venv, required = TRUE)
py_install('scanorama', method = 'virtualenv')
After this, you can install additional python packages in this virtual environment by calling only py_install.
Jupyter notebook
With the jupyter notebook interface you can do most of the python-based analyses (e.g. based on python notebook (.ipynb) files). The software is installed in a conda environment. The environment for the general exercises is called scRNASeq2021. You can use this environment in a python notebook by selecting the right kernel. You can add installations to the environment by using the terminal (e.g. with conda install mypackage). You can also create new environments.
Download data using S3
There are two ways to download data from the backup at s3:
- Using
aws-cli - Using
wgetorcurl
Download with AWS command line interface
Install aws-cli, e.g. with conda:
conda install -c conda-forge awscli
To use the AWS command line interface you will need an AWS account and set up awscli. Follow instructions here.
To download your home directory use the following command:
aws s3 cp s3://single-cell-summerschool/general/users/[USERNAME]/ ./homedir --recursive
Replace [USERNAME] with your username (e.g. gvangeest). You can find back your username in the google spreadsheat.
Note: Participants of the deep learning project should replace general in the path with gpu2.
Download with wget or curl
You can download specific files without aws-cli. To do this, use the following command:
wget https://single-cell-summerschool.s3.eu-central-1.amazonaws.com/general/users/[USERNAME]/[FILENAME]
or curl:
curl -O https://single-cell-summerschool.s3.eu-central-1.amazonaws.com/general/users/[USERNAME]/[FILENAME]
Replace [FILENAME] with the exact filename you want to download, and replace [USERNAME] with your username (e.g. gvangeest). You can find back your username in the google spreadsheat.
 Conda
Conda
During this workshop, you can use conda environments to run the exercises (see AWS instance if you want to use the cloud-based solution). This is because conda environments allow all users to have the same computing environment, i.e. package versions. This enforces reproducibility for you to run this material without the need to re-install or change your local versions. See and graphical example below:
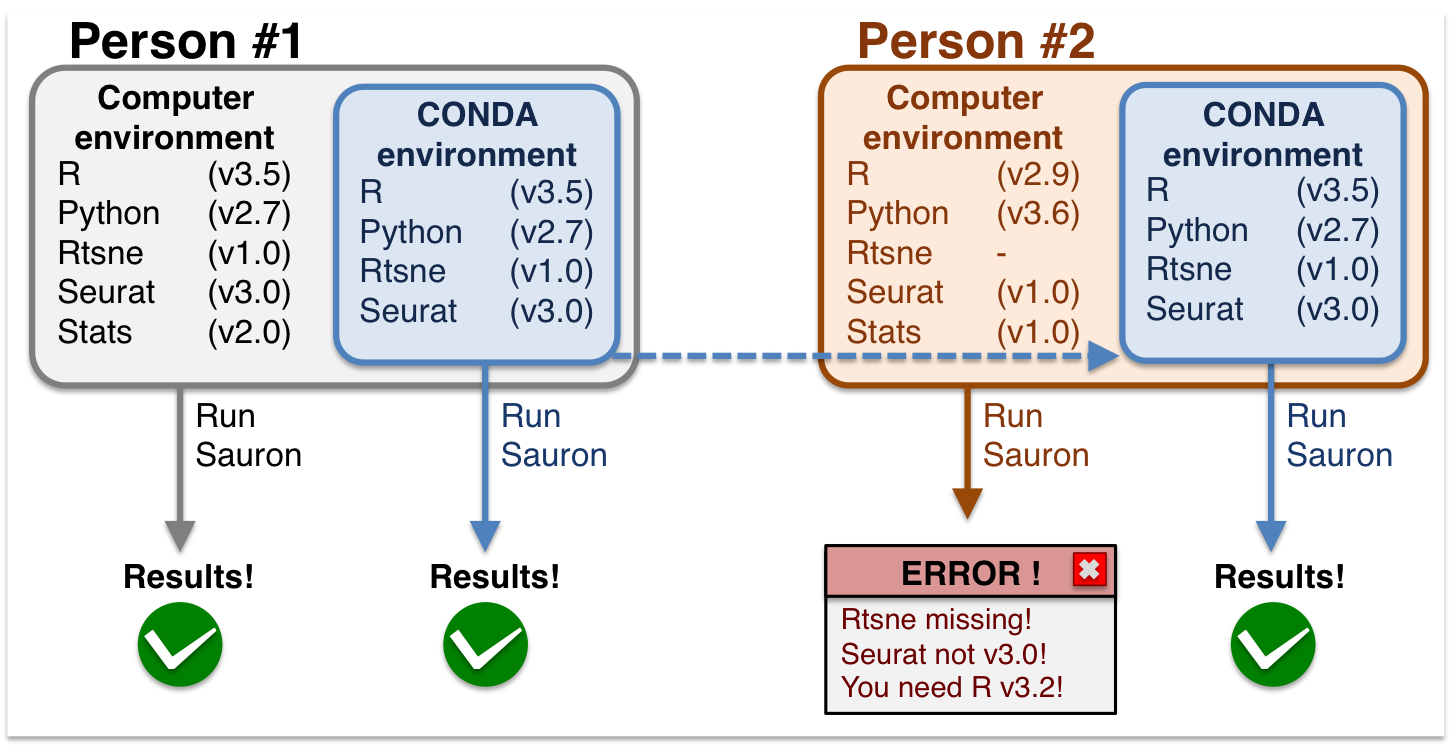
Conda environments are a self-contained directory that you can use in order to reproduce all your results.
Briefly, you need to:
- Install Conda
- Download the
.ymlenvironment file - Create and activate the environment
- Deactivate the environment after running your analyses
You can read more about Conda environments and other important concepts to help you make your research reproducible.
1. Download and install Conda and Mamba
Start by installing Conda. We suggest installing Miniconda3 and NOT Anaconda. After installing Conda.
On Mac OS X
First, make sure you have Xcode and CommandLineTools installed and updated to latest version (in AppStore). If you have not already installed CommadLineTools, go to a terminal window and run:
xcode-select --install
First download the latest version of Miniconda3 and run it to install.
curl -o Miniconda3-latest-MacOSX-x86_64.sh https://repo.anaconda.com/miniconda/Miniconda3-latest-MacOSX-x86_64.sh
sh Miniconda3-latest-MacOSX-x86_64.sh
Follow the instructions on screen, scrolling down, pressing ENTER and replying yes when necessary. Install it in the default directory. Restart your terminal window to apply modifications. After restarting, you can type the command below to install Mamba:
conda init
conda install -n base -c conda-forge mamba
On Ubuntu
First download the latest version of Miniconda3 and run it to install.
wget https://repo.anaconda.com/miniconda/Miniconda3-latest-Linux-x86_64.sh
sh Miniconda3-latest-Linux-x86_64.sh
Follow the instructions on screen replying yes when necessary. Restart your terminal window to apply modifications. After restarting, you can type the command below to install Mamba:
conda init
conda install -n base -c conda-forge mamba
On Windows 10
Unfortunately, not all packages available on conda are compatible with windows machines. The good news is that Windows 10 offers native linux support via the Windows Subsystem for Linux (WSL2). This allows you to run linux/bash commands from within windows without the need of a virtual machine nor a dual-boot setup (i.e. having 2 operating systems). However, WSL does not offer a complete support for graphical interfaces (such as RStudio in our case), so we need additional steps to make that happen.
-
On Windows 10, install the WSL if you don't have it. Follow the instructions here: https://docs.microsoft.com/en-us/windows/wsl/install-win10
-
Once you have that installed, you can download and install MobaXterm (which is the enhanced terminal with graphical capacity): https://mobaxterm.mobatek.net
It is recommended that you INSTALL the program and not use the portable version. -
Inside MobaXterm, you will probably will see that your WSL is already listed on the left panel as an available connection. Just double-click it and you will be accessing it via MobaXterm. If by any chance you don't see it there, close MobaXterm and go to the WSL terminal, because probably the WSL is not allowing SSH connections. You can follow this link for the instructions on how to do it. You need to complete until the step
Start or restart the SSH service, while the further steps are optional, but might be useful. -
Inside MobaXterm, download Conda with the command:
wget https://repo.anaconda.com/miniconda/Miniconda3-latest-Linux-x86_64.sh
- Inside MobaXterm, type the commands below to install Conda. Follow the instructions for the installation there.
cd ~/Downloads
sh Miniconda3-latest-Linux-x86_64.sh
- Inside MobaXterm, Follow the instructions on screen replying
yeswhen necessary. Restart your terminal window to apply modifications. After restarting, you can type the command below to install Mamba:
conda init
conda install -n base -c conda-forge mamba
- Inside MobaXterm, type the commands below to install the X-server graphical packages that will be used to launch RStudio. https://docs.anaconda.com/anaconda/install/linux/
sudo apt-get update
sudo apt-get install libgl1-mesa-glx libegl1-mesa libxrandr2 libxrandr2 libxss1 libxcursor1 libxcomposite1 libasound2 libxi6 libxtst6
- Close and open all application and Inside MobaXterm, you will probably will see that your WSL is already listed on the left panel as an available connection. Just double-click it and you will be accessing it via MobaXterm.
On VirtualBox
If by any means you see that the installations are not working as it should on your computer, you can try to create a virtual machine to run UBUNTU and install everything there. But please keep this alternative as the last temporary resourse, as we recommend troubleshooting the installation o the up-mentioned methods.
-
Download and install on your machine VIRTUALBOX https://www.virtualbox.org
-
Download the ISO disk of UBUNTU https://ubuntu.com/download/desktop
-
On VIRTUALBOX, click on
Settings(yellow engine) >General>Advancedand make sure that both settings Shared Clipboard and Drag'n'Drop are set toBidirectional. -
Completely close VIRTUALBOX and start it again to apply changes.
-
On VIRTUALBOX, create a machine called Ubuntu and add the image above
- set the memory to the maximum allowed in the GREEN bar
- set the hard disk to be dynamic allocated
- all other things can be default
-
Proceed with the Ubuntu installation as recommended. You can set to do "Minimal Installation" and deactivate to get updates during installation.
-
Inside Ubuntu, open TERMINAL and type the commands below to install the X-server graphical packages that will be used to launch RStudio. https://docs.anaconda.com/anaconda/install/linux/
sudo apt-get update
sudo apt-get install libgl1-mesa-glx libegl1-mesa libxrandr2 libxrandr2 libxss1 libxcursor1 libxcomposite1 libasound2 libxi6 libxtst6
- Inside UBUNTU, Download conda:
wget https://repo.anaconda.com/miniconda/Miniconda3-latest-Linux-x86_64.sh
- Inside UBUNTU, open the TERMINAL and type the commands below. Follow the instructions for the installation there.
cd ~/Downloads
sh Miniconda3-latest-Linux-x86_64.sh
- Close Terminal to apply the CONDA updates. Then you can create a course folder, download the environment file and create the environment:
2. Create a conda environment from file
To download the environment_scRNAseq2021.yml file using the command on Terminal:
curl -o environment_scRNAseq2021.yml https://raw.githubusercontent.com/NBISweden/single-cell_sib_scilifelab_2021/main/environment_scRNAseq2021.yml
After this, you should have a file named environment_scRNAseq2021.yml in your directory (it does not matter where). Next, type:
mamba env create -n scRNAseq2021 -f environment_scRNAseq2021.yml
Several messages will show up on your screen and will tell you about the installation process. This may take a few minutes depending on how many packages are to be installed.
##Collecting package metadata: done
##Solving environment: done
##
##Downloading and Extracting Packages
##libcblas-3.8.0 | 6 KB | ############################################################################# | 100%
##liblapack-3.8.0 | 6 KB | ############################################################################# | 100%
##...
##Preparing transaction: done
##Verifying transaction: done
##Executing transaction: done
3. Activate the environment
Once the environment is created, we need to activate it in order to use the softwares and packages inside it. To activate an environment type:
conda activate scRNAseq2021
From this point on you can run any of the contents from the workshop. For instance, you can directly launch RStudio by typing rstudio. Here it is important to add the & symbol in the end to be able to use the command line at the same time if needed. You can open other files from Rstudio later as well.
rstudio PATH/my_script.Rmd &
4. Deactivate the environment
After you've ran all your analyses, you can deactivate the environment by typing:
conda deactivate
 Dataset
Dataset
TBA
 Code
Code
TBA