Putting it all together
26-Jan-2026
Working reproducibly will make your research life a lot easier!

Take control of your research by making its different components reproducible
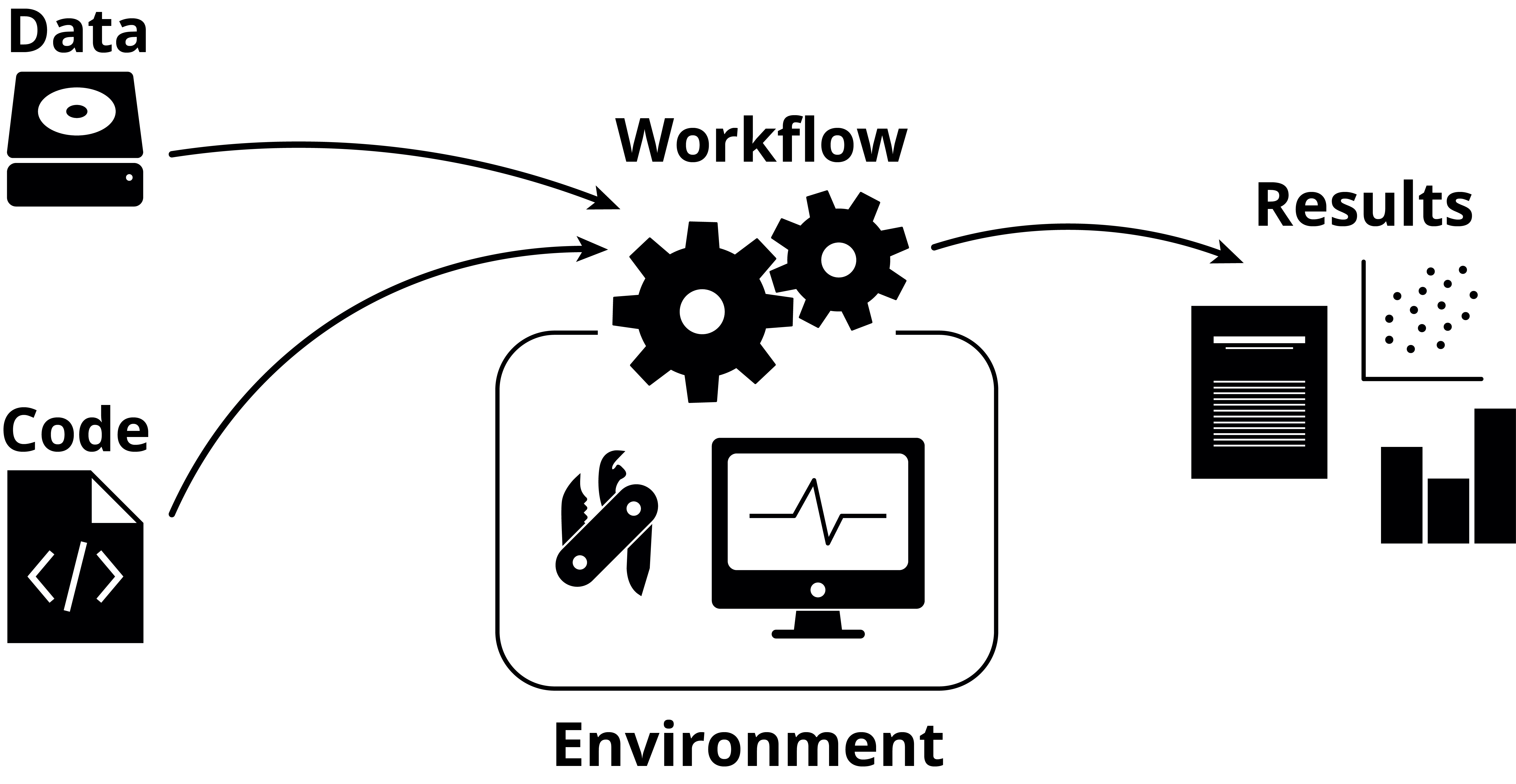
What have we learned?

- How to use the version control system Git to track changes to code
- How to use the package and environment manager Conda
- How to use the workflow managers Snakemake and Nextflow
- How to use Quarto and Jupyter to generate automated reports and to document your analyses
- How to use Docker and Apptainer to distribute containerized computational environments
- How to create and maintain proper documentation
Divide your work into distinct projects
- Keep all files needed to go from raw data to final results in a dedicated directory
- Use relevant subdirectories
- Use Git to version control your projects
- Do not store data and results/output in your Git repository
- When in doubt, commit often rather than not
Find your own project structure
For example:
code/ Code needed to go from input files to final results
data/ Raw data - this should never edited
doc/ Documentation of the project
env/ Environment-related files, e.g. Conda environments or Dockerfiles
results/ Output from workflows and analyses
README.md Project documentationMore examples:
Treasure your data
- Keep your raw data read-only and static
- Don’t create different versions of the input data - write a script, Quarto document, Jupyter notebook or a Snakemake / Nextflow workflow if you need to pre-process your input data so that the steps can be recreated
- Backup! Keep redundant copies in different physical locations
- Upload your raw data as soon as possible to a public data repository
Organise your coding
- Avoid generating files interactively or doing things by hand
- Write scripts, Quarto documents, Jupyter notebooks or Snakemake / Nextflow workflows for reproducible results to connect raw data to final results
- Keep the parameters separate (e.g. at top of file or in a separate configuration file)
What is reasonable for your project?

What is reasonable for your project?
Minimal
Write code in a reproducible way and track your environment
- Track your projects with a Git repository each; publish code with your results on e.g. GitHub
- Use Conda to install software in environments that can be exported and installed on a different system
- Publish your
environment.ymlfile along with your code
What is reasonable for your project?
Good
Structure and document your code with notebooks
- Use Quarto or Jupyter notebooks to better keep track of and document your code
- Track your notebooks with Git
What is reasonable for your project?
Great
Track the full environment and connect your code in a workflow
- Go one step beyond in tracking your environment using Docker or Apptainer
- Convert your code into a Snakemake / Nextflow workflow
- Track both your image definitions (e.g. Dockerfiles) as well as your workflows with Git
Alternatives
Version control
- Git – Widely used and a lot of tools available + GitHub/BitBucket.
- Jujutsu - New “front-end” for Git by Google, with a smaller and improved mental model for version control.
- Mercurial – Distributed model just like Git, close to Sourceforge.
- Subversion – Centralized model unlike git/mercurial; no local repository on your computer and somewhat easier to use.
Alternatives
Environment / package managers
- Conda – General purpose environment and package manager. Community-hosted collections of tools at Bioconda or Conda-forge.
- Pixi - General purpose environment/package manager built on the Conda ecosystem, but much faster and works seamlessly with lock-files.
- Pip – Package manager for Python, has a large repository at PyPI.
- Apt/yum/brew – Native package managers for different OS. Integrated in OS and might deal with e.g. update notifications better.
- Virtualenv – Environment manager used to set up semi-isolated python environments.
Alternatives
Workflow managers
- Snakemake – Based on Python, easily understandable format, relies on file names.
- Nextflow – Based on Groovy, uses data pipes rather than file names to construct the workflow.
- Make – Used in software development and has been around since the 70s. Flexible but notoriously obscure syntax.
- Galaxy - attempts to make computational biology accessible to researchers without programming experience by using a GUI.
Alternatives
Literate programming
- Quarto - Developed by Posit (previously RStudio), command-line tool focused on generating high-quality documents in a language-agnostic way.
- Jupyter – Interactive notebooks in a variety of languages/formats.
- Marimo - Similar to Jupyter, but pure python. Tries to eliminate common problems with Jupyter notebooks.
- R Markdown – Developed by Posit (previously RStudio), focused on generating high-quality documents.
- Zeppelin – Developed by Apache. Closely integrated with Spark for distributed computing and Big Data applications.
- Beaker – Newcomer based on IPython, just as Jupyter. Has a focus on integrating multiple languages in the same notebook.
Alternatives
Containerization / virtualization
- Docker – Used for packaging and isolating applications in containers. Dockerhub allows for convenient sharing. Requires root access.
- Apptainer/Singularity – Simpler Docker alternative geared towards high performance computing. Does not require root.
- Podman - open source daemonless container tool similar to docker in many regards
- Shifter – Similar ambition as Singularity, but less focus on mobility and more on resource management.
- VirtualBox/VMWare – Virtualisation rather than containerization. Less lightweight, but no reliance on host kernel.
“What’s in it for me?”

NBIS Bioinformatics drop-in
Any questions related to reproducible research tools and concepts? Talk to an NBIS expert!
- Online Drop-in: https://meet.nbis.se/dropin
- Every Tuesday, 14.00-15.00 (except public holidays)
- Check https://www.nbis.se/get-support/talk-to-us for more info

