Making reproducible workflows with 
26-Jan-2026
Nextflow features
- Generalisable
- Portable
- Scalable
- Platform-agnostic
- Based on Groovy and Java
- Large active community in e.g. nf-core
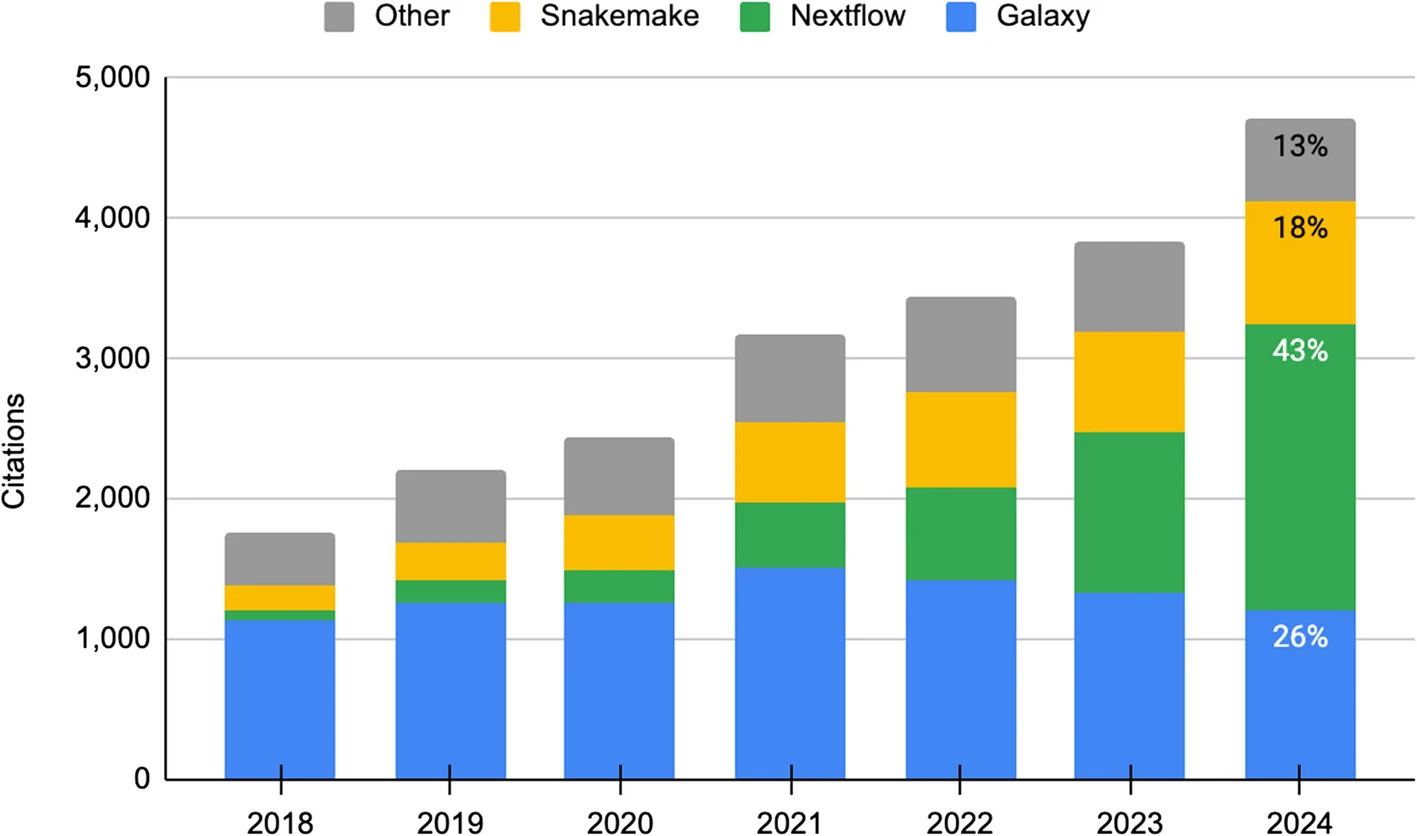
Nextflow adoption
Nextflow has grown rapidly since its creation, and is the most commonly used workflow manager in bioinformatics since 2024. 1
Concepts and nomenclature
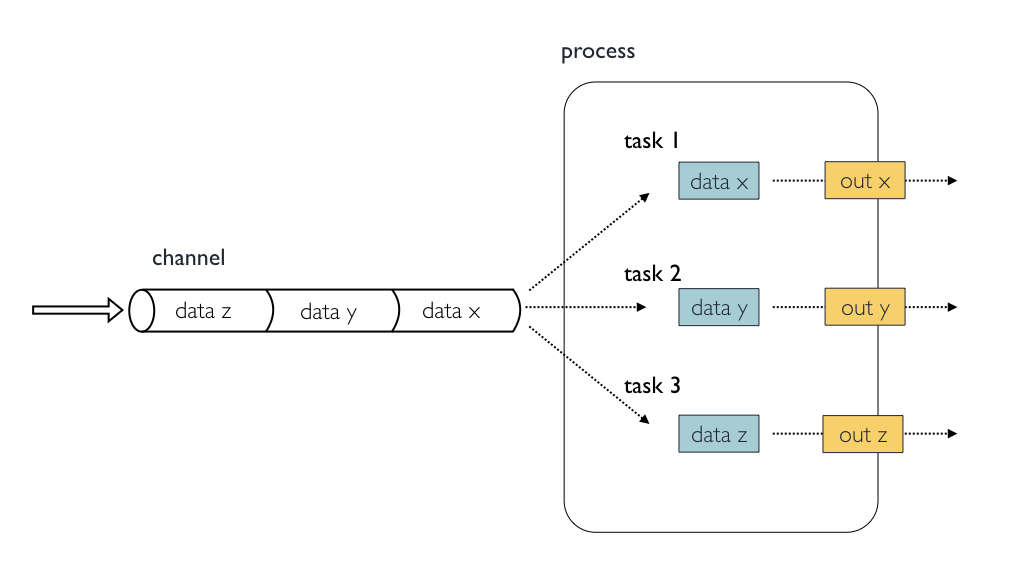
- Channels contain data, e.g. input files
- Processes run some kind of code, e.g. a script or a program
- Tasks are instances of a process, one per process input
- Each task is run in its own, isolated sub-directory
Anatomy of a process
Anatomy of a process
Anatomy of a process
Anatomy of a process
Anatomy of a workflow
Anatomy of a workflow
Anatomy of a workflow
Executing Nextflow
Snakemake and Nextflow differences
| Snakemake | Nextflow | |
|---|---|---|
| Language | Python | Groovy |
| Data | Everything is a file | Can use both files and values |
| Execution | Working directory | Each job in its own directory |
| Philosophy | “Pull” | “Push” |
| Dry-runs | Yes | No |
- Question: But, which one is the best?
- Answer: Both - it’s mostly up to personal preference!

