Tools for Reproducible Research
Introduction
26-Jan-2026
Course content
Good practices for working with data
How to use the version control system Git to track changes to code
How to use the package and environment manager Conda
How to use the workflow managers Snakemake and Nextflow
How to generate automated reports using Quarto and Jupyter
How to use Docker and Apptainer to distribute containerised computational environments
The Teachers







What is NBIS?
National Bioinformatics Infrastructure Sweden
- A distributed national bioinformatics infrastructure supporting life sciences in Sweden
- Provides hands-on bioinformatic support, training, infrastructure and a weekly drop-in
- Situated throughout Sweden
- Provides wide-spectrum support in the fields of bioinformatics, bioimage informatics, data management, imaging AI, development of systems and tools as well as national compute resources.
- Read more at nbis.se
What is reproducibility?
Why all the talk about reproducibility?
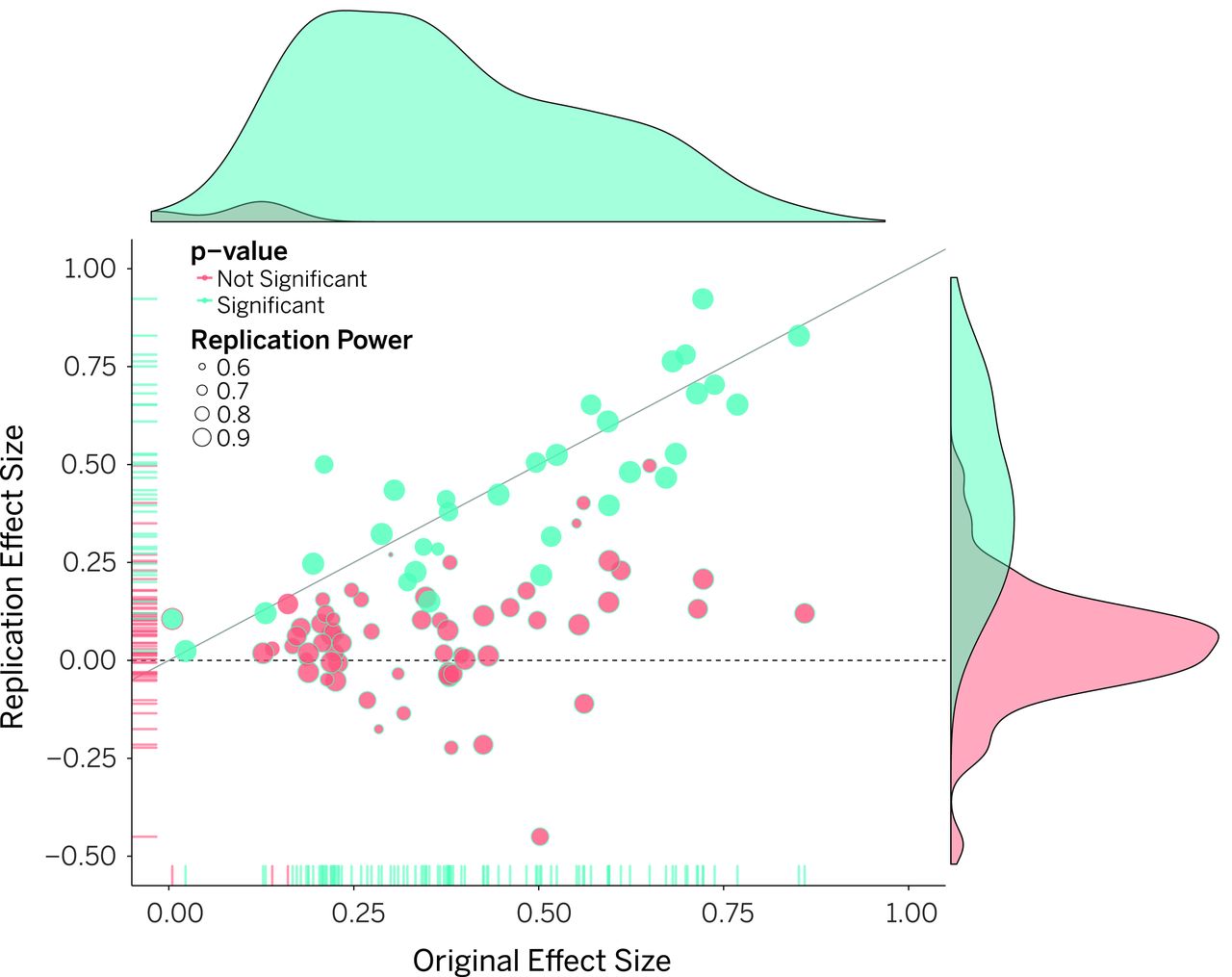
In 2015 the Open Science Collaboration set out to replicate 100 experiments published in high-impact psychology journals. 1
- Less than 50% of the experiments could be replicated
- Effect sizes were significantly smaller in the replicated studies
Why all the talk about reproducibility?
The same year, money spent on preclinical research that could not be reproduced was estimated at $28 billion in the US. 1
Why all the talk about reproducibility?
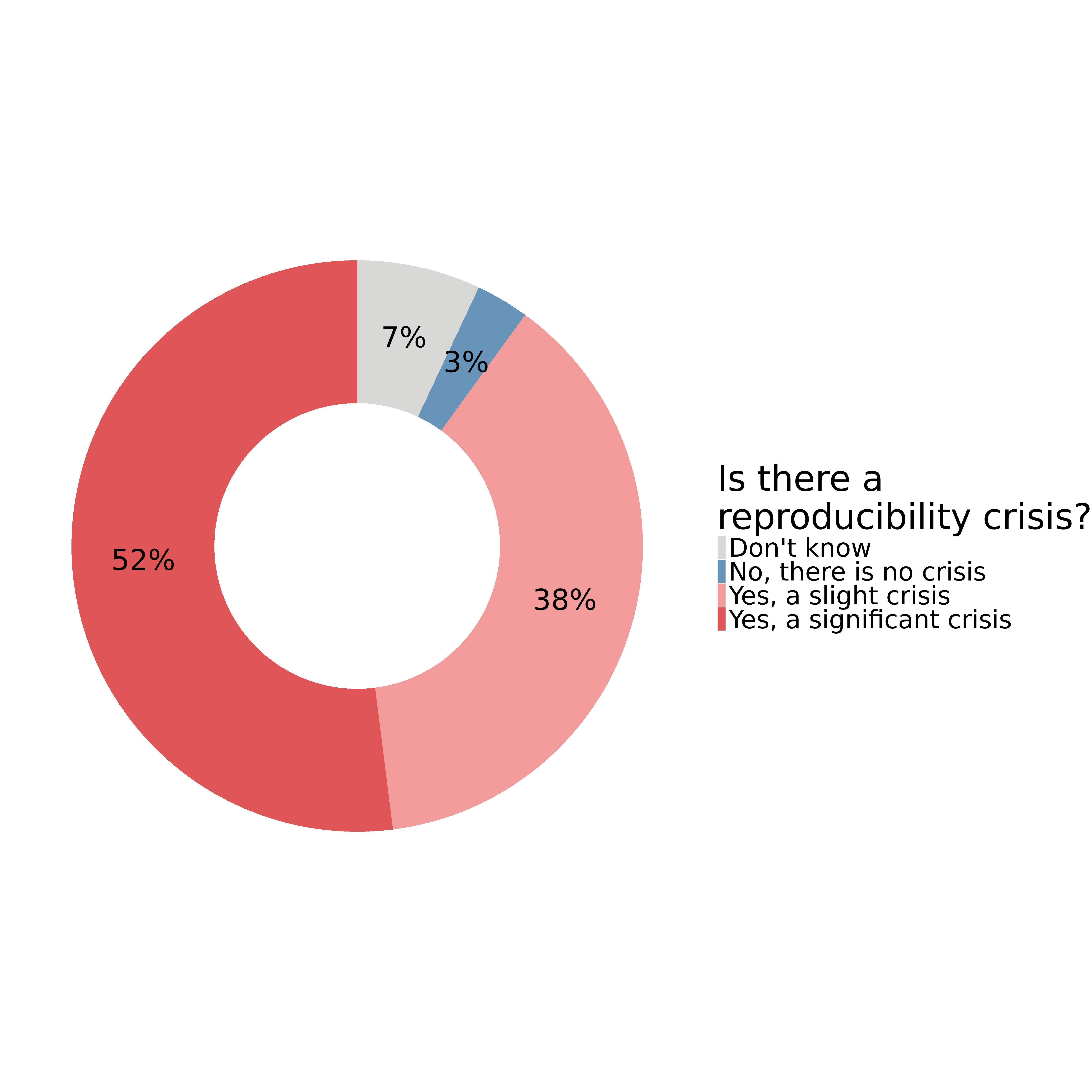
In 2016, 1,576 scientists were surveyed about reproducibility. 1
- 90% agreed that there is a ‘slight’ or ‘significant’ reproducibility crisis
Why all the talk about reproducibility?
In 2016, 1,576 scientists were surveyed about reproducibility. 1
- 90% agreed that there is a ‘slight’ or ‘significant’ reproducibility crisis
- Failure to reproduce experiments is a problem across all domains of science
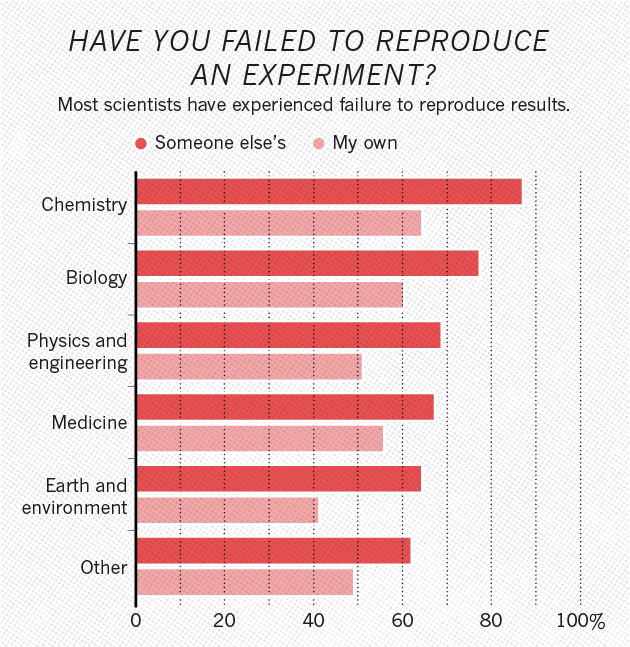
Reproducibility in computational research
In 2018, Stodden et al estimated the reproducibility rate of computational papers published in the journal Science. 1
- Only 26% of the studies were estimated to be reproducible
- Failure to reproduce was mainly due to lack of data and code
- Stricter journal guidelines gave improvement but were insufficient
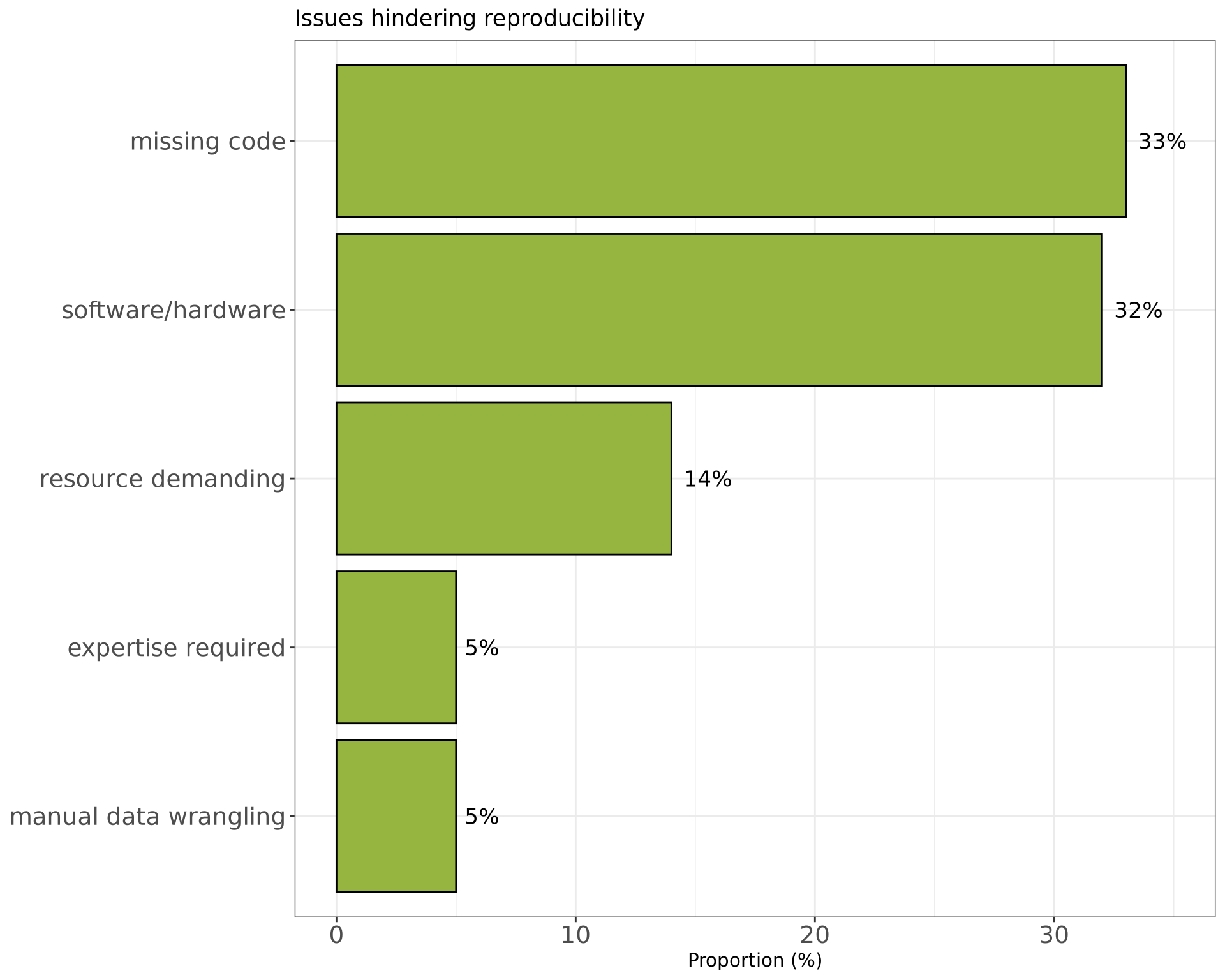
Reproducibility in computational research
More examples:
- Trisovic et al (2022) found that only 26% of R files in the Harvard Dataverse could be executed as-is.1
- Sheeba & Mietchen (2024) found that only 8% of Jupyter notebooks used in publications executed without errors.2
- Missing dependencies
- Missing variables
- Incorrect file/directory structure
Implications for research
Innovation points out paths that are possible; replication points out paths that are likely; progress relies on both. 1
What does reproducible research mean?
| Data | |||
| Same | Different | ||
| Code | Same | Reproducible | Replicable |
| Different | Robust | Generalisable | |
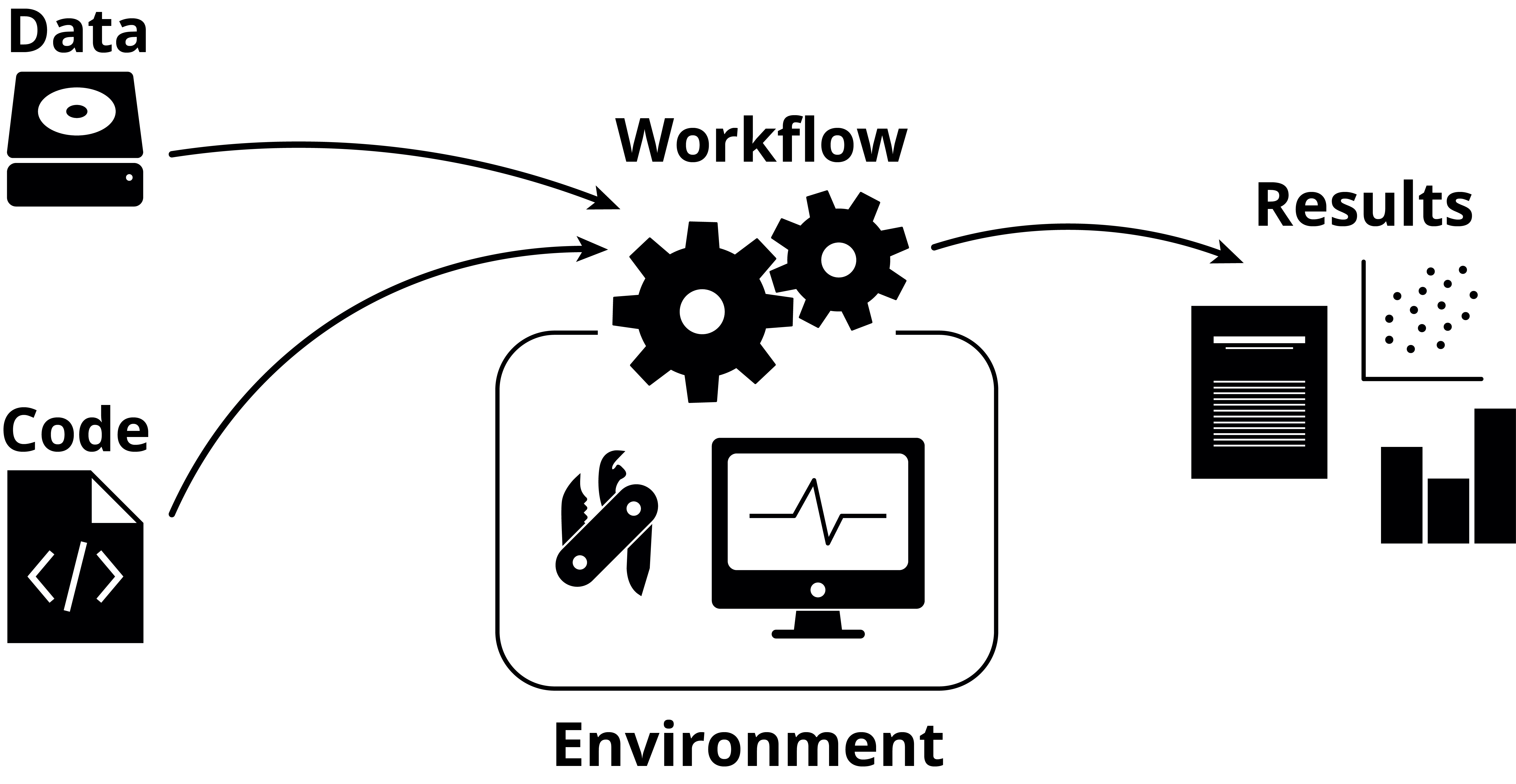
How are you handling your data?
Decent:
- Data available on request
- All metadata required for generating the results available
Good:
- Raw data deposited in public repositories
- If the raw data needed preprocessing, scripts were used rather than modifying it manually
Great:
- Section in the paper or online repository (e.g. GitHub) to aid in reproduction
- Used non-proprietary and machine-readable formats, e.g.
.csvrather than.xls.
How are you handling your code?
Decent:
- All code for generating results from processed data available on request
Good:
- All code for generating results from raw data is available
- The code is publicly available with timestamps or tags
Great:
- Code is documented and contains instructions for reproducing results
- Seeds were used and documented for heuristic methods
How are you handling your environment?
Decent:
- Key programs used are mentioned in the materials and methods section
Good:
- List of all programs used and their respective versions are available
Great:
- Instructions for reproducing the whole environment publicly available
“What’s in it for me?”
Before the project:
- Improved structure and organisation
- Forced to think about scope and limitations
During the project:
- Easier to re-run analyses and generate results after updates and/or changes
- Closer interaction between collaborators
- Much of the manuscript “writes itself”
After the project:
- Faster resumption of research by others (or, more likely, your future self)
- Increased visibility in the scientific community

