Schematic workflow
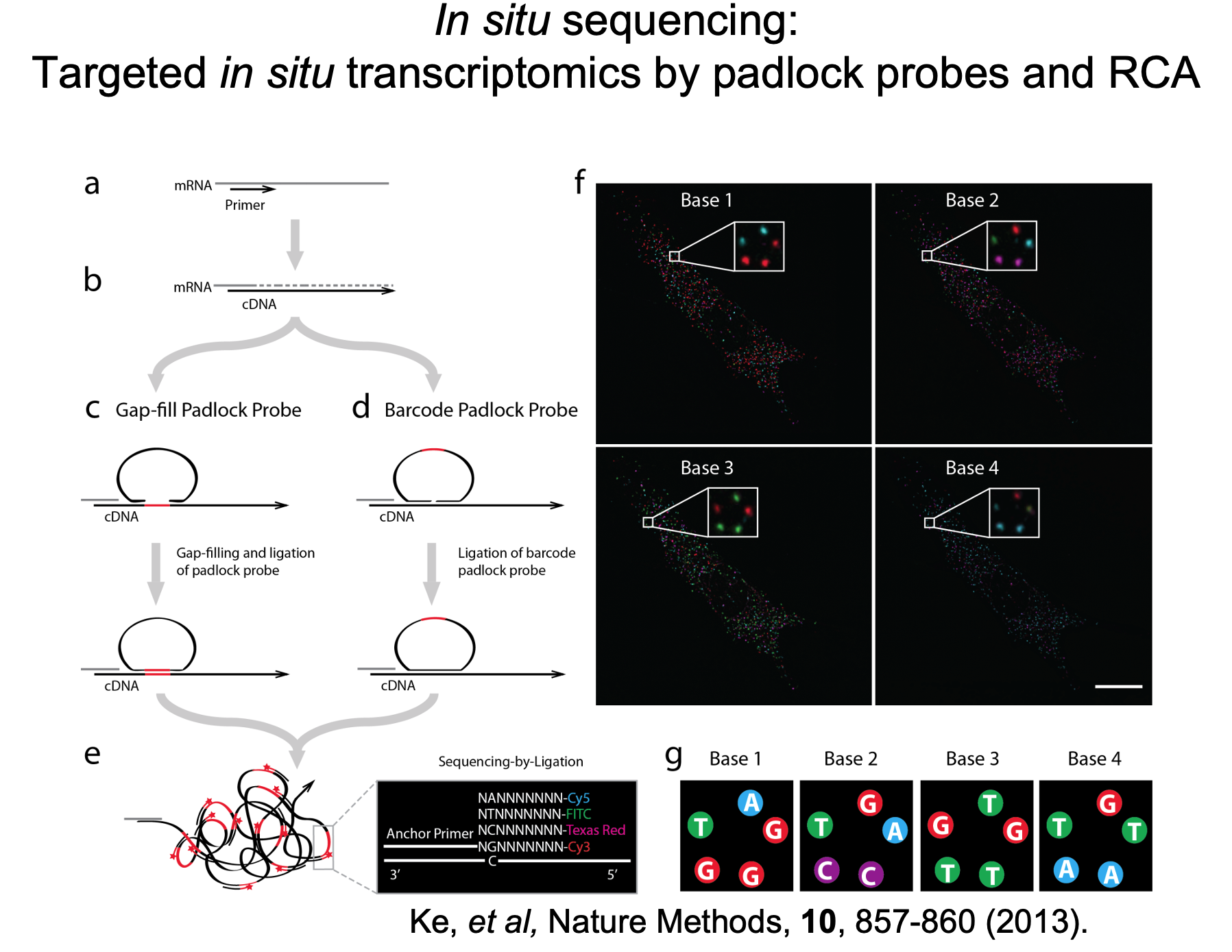
General information
The In Situ Sequencing (ISS) unit offers spatially resolved gene expression analysis for targeted panels of genes at subcellular resolution. Developed in the lab of Mats Nilsson, a pioneer in generating in situ gene expression and mutation profiles (Ke R et al., 2013, Nature Methods), ISS provides a cutting-edge approach to spatial transcriptomics.
Key Features of ISS:
- Preservation of Spatial Context: Maintains the spatial organization of gene expression within the tissue architecture.
- Targeted Approach: Utilizes padlock probes for precise gene targeting. High Specificity: Employs advanced amplification methods for enhanced molecular specificity.
- Multiplexing Capability: Detects up to several hundred transcripts per sample. Adjustable Sensitivity: Sensitivity can be fine-tuned to match experimental needs. High Throughput: Achieved through wide-field imaging technology.
ISS enables simultaneous localization and quantification of over 100 transcripts with subcellular resolution in a single tissue section, providing an unparalleled view of gene expression in its native spatial context.
Workflow and Integration
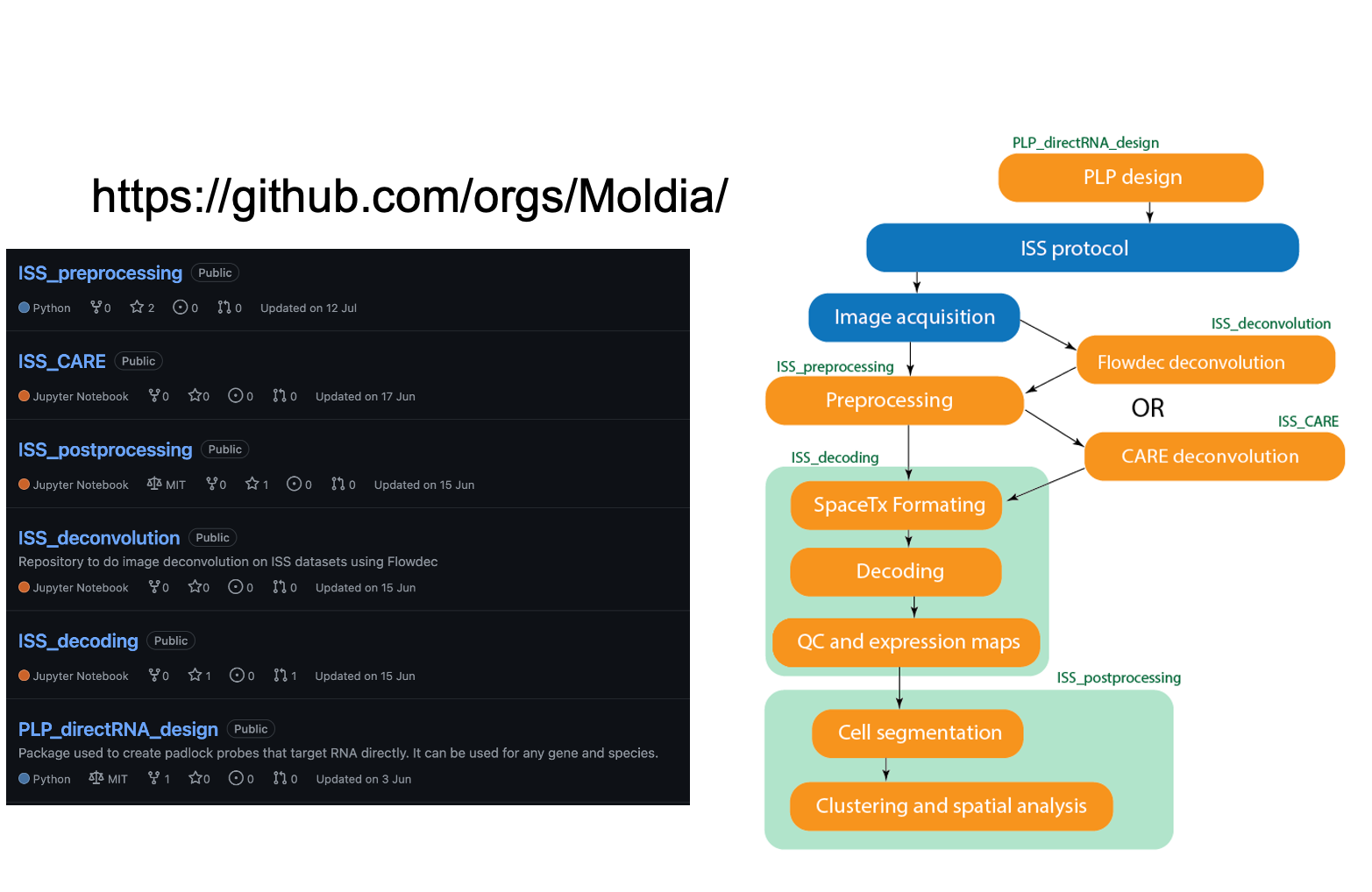
In house ISS workflow
In house ISS workflow is developed to analyse data generated by in house ISS technique.
 :
:
Niche Identification
NicheCompass: This workflow, developed by NBIS, is a nextflow pipeline which runs NicheCompass which is a package for end-to-end analysis of spatial multi-omics data, including spatial atlas building, niche identification & characterization, cell-cell communication inference and spatial reference mapping.
PLP probe design
In this portal, you can design your probes of interest for in situ seuqencing, develeoped by NBIS.