| len | supp | dose |
|---|---|---|
| 4.2 | VC | 0.5 |
| 11.5 | VC | 0.5 |
| 7.3 | VC | 0.5 |
| 5.8 | VC | 0.5 |
| 6.4 | VC | 0.5 |
| 10.0 | VC | 0.5 |
R packages
RaukR 2024 • Advanced R for Bioinformatics
Sebastian DiLorenzo
21-Jun-2024
Overview
- What is an R package?
- Possible package states
- Package structure:
- Code |
r/ - Metadata |
DESCRIPTION - Documentation |
man/ - Vignettes
- Import & Export |
NAMESPACE - Data |
data/ - Tests |
tests/ - Compiled code |
src/
- Code |
- CRAN and
R CMD check - Rstudio and Github
What is an R package?
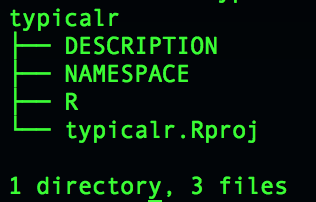
- A strict and connected folder and file structure
What is an R package?

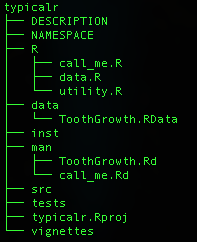
- A strict and connected folder and file structure
What is an R package?


- A strict and connected folder and file structure
- Sharing code
- Improved quality and rigor
- Documentation
- Tests
- Examples
- Efficiency
- Improvability
- Reproducibility
Package naming
- A name that describes your packages function
- Letters, numbers and periods
- Must start with letter
- Cannot end with period
- Make it “googleable”
- Check that it doesn’t already exist!
- CRAN
- GitHub
- Bioconductor
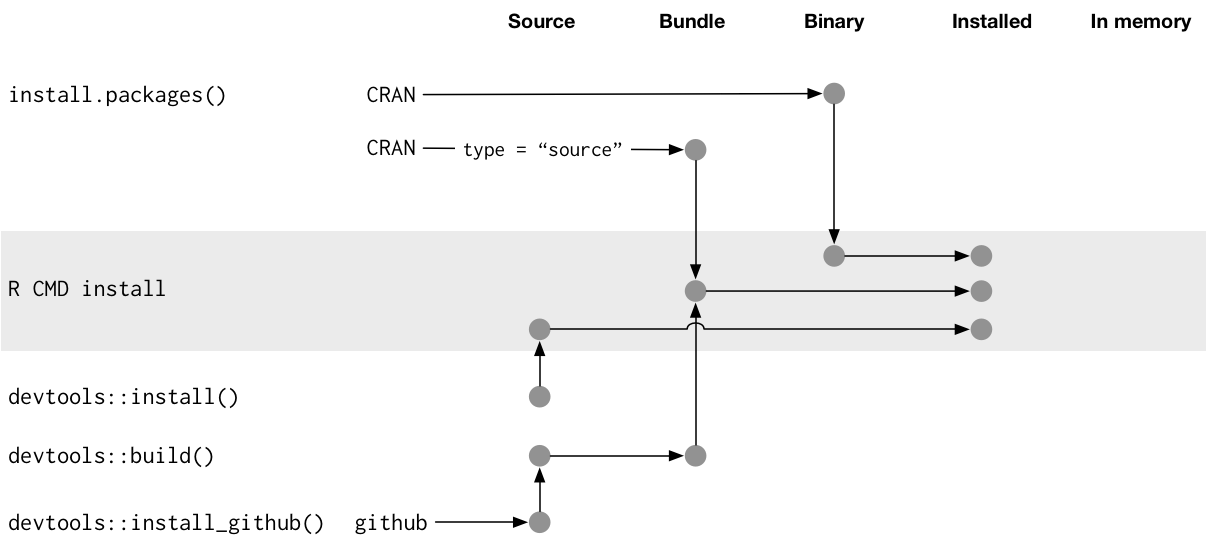
Package states
There are five states a package can exist in:
- Source
- Bundled
- Binary
- Installed
- In-memory
Package states
Source
The development version of your package. The collection of files on your computer.
Bundled
- A compressed, tar.gz source package with vignettes built
- .Rbuildignore files are excluded
- Useful for data for example
Binary
- A bundle that is built for a certain architecture
- Parsed format, skipping the development tools needed to take the package between source and being interpretable by R
Package states
Installed
- A binary package decompressed into a package library for R
- The package library is the directory or directories where
library(packagename)searches.libPaths()
- The package library is the directory or directories where
In-memory
When you use a package, it is in memory. When developing, a package does not have to be installed to be in memory.
packagename::function()loads packagenamelibrary(packagename)loads and attaches packagename
Package states
R/

R/

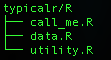
- Code
- Large functions in their own R files
- Utility functions, that your package uses, in one R file
- Bad code
library(),require(),source()options(),par()
DESCRIPTION
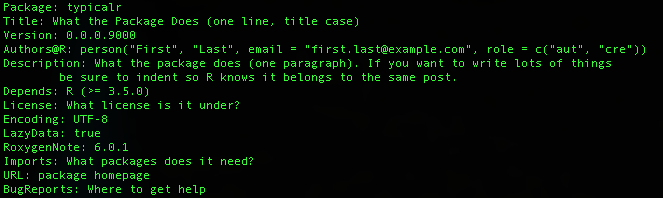
- Title
- 65 characters, no punctuation
- Version
- The version of the package
- Description
- One paragraph
- Authors@R
- Roles
- cre*: Creator or maintainer.
- aut*: Author or authors, that have made significant contributions.
- ctb: Contributors, have made smaller contributions.
- cph: Copyright holder. Used if copyright is held by someone other than author, typically company.
- Roles
DESCRIPTION

- Depends & Imports
- Packages and versions that your package needs
- Versions are optional
- Depends: Attaches!
- Imports: Loads!
- Suggests
- Added functionality
- LazyData
- Datasets occupy no memory until loaded
- License
- Can be a file; LICENSE
- Influences permissions of who can distribute and modify in what way
- Most common; MIT, GPL-3, CC0.
- https://tldrlegal.com/
- CRAN requires a license
Versioning
0.0.0.9000
major.minor.patch.dev
- Major
- Large changes, not always backwards compatible
- Usually 1 upon first release out of dev
- Minor
- Bug fixes & new features. Most common
- Patch
- Small bugfixes, no new features.
- Dev
- Only used while under development
- Always starts at 9000
man/

call_me.R
#' Output "Call me " followed by input.
#'
#' @param x A character or characters.
#' @return The string "Call me " and \code{x}. I'll write this
#' to display how to section with tags.
#' @examples
#' call_me("Maeby")
call_me <- function(x) {
paste("Call me ", x, sep="")
}Roxygen2- ?function
- Comment block,
#', preceding a function - Tags,
@tags, map values - No tag for introduction
- title*
- description
- details
- Special characters
@\%, escape with\
man/
call_me.R
#' Output "Call me " followed by input.
#'
#' @param x A character or characters.
#' @return The string "Call me " and \code{x}. I'll write this
#' to display how to section with tags.
#' @examples
#' call_me("Maeby")
call_me <- function(x) {
paste("Call me ", x, sep="")
}call_me.Rd
% Generated by roxygen2: do not edit by hand
% Please edit documentation in R/call_me.R
\name{call_me}
\alias{call_me}
\title{Output "Call me " followed by input.}
\usage{
call_me(x)
}
\arguments{
\item{x}{A character or characters.}
}
\value{
The string "Call me " and \code{x}. I'll write this
to display how to section with tags.
}
\description{
Output "Call me " followed by input.
}
\examples{
call_me("Maeby")
}]{.small}
> ?call_me

man/ for datasets

data.R:
#' The Effect of Vitamin C on Tooth Growth in Guinea Pigs
#'
#' The response is the length of odontoblasts (cells responsible for tooth growth)
#' in 60 guinea pigs. Each animal received one of three dose levels of vitamin C
#' (0.5, 1, and 2 mg/day) by one of two delivery methods, orange juice or ascorbic
#' acid (a form of vitamin C and coded as VC).
#'
#' @usage ToothGrowth
#'
#' @format A data frame with 60 observations on 3 variables.
#' \describe{
#' \item{len}{Tooth length}
#' \item{supp}{Supplement type (VC or OJ).}
#' \item{dose}{Dose in milligrams/day}
#' }
#' @source \url{https://www.elsevier.com/books/the-statistics-of-bioassay/bliss/978-1-4832-5662-7}
"ToothGrowth"vignettes/
- A more complete guide to your package
- For user/you
- Examples and use cases
knitr&rmarkdownknitr: add r code to markdown
vignettes/package-vignette.Rmd
usethis::use_vignette("typicalr-vignette")typicalr-vignette.Rmd
---
title: "Vignette Title"
author: "Vignette Author"
date: "2024-05-29"
output: rmarkdown::html_vignette
vignette: >
%\VignetteIndexEntry{Vignette Title}
%\VignetteEngine{knitr::rmarkdown}
%\VignetteEncoding{UTF-8}
---vignettes/
- A more complete guide to your package
- For user/you
- Examples and use cases
knitr&rmarkdownknitr: add r code to markdown
vignettes/package-vignette.Rmd
usethis::use_vignette("typicalr-vignette")typicalr-vignette.Rmd
---
title: "typicalr"
author: "Sebastian DiLorenzo"
date: "2024-05-29"
output: rmarkdown::html_vignette
vignette: >
%\VignetteIndexEntry{typicalr}
%\VignetteEngine{knitr::rmarkdown}
%\VignetteEncoding{UTF-8}
---NAMESPACE
package1 names package2 names
@importsand@importsFrom- Defines how/where a function in one package finds a function in another
@importspkg@importsFrompkg function
@export- Defines which functions are available to user
- Do not export data
NAMESPACE
call_me.R:
#' Output "Call me " followed by input.
#'
#' @param x A character or characters.
#' @return The string "Call me " and \code{x}. I'll write this
#' to display how to section with tags.
#' @examples
#' call_me("Maeby")
#' @export
call_me <- function(x) {
paste("Call me ",x,sep="")
}utility.R:
#' @import knitr
NULLNAMESPACE:
# Generated by roxygen2: do not edit by hand
export(call_me)
import(knitr)NAMESPACE
Import in DESCRIPTION and in NAMESPACE!?
- DESCRIPTION
Imports:- “My package needs this package to work”
- NAMESPACE
@import- “When my package uses this function, use the one from the package in the NAMESPACE”
- Additional effects:
- NAMESPACE removes need for
::package::function()orfunction()
- NAMESPACE removes need for
data/
Package types:
- Functional
- Performs a or several functions
- Contains no or small datasets, <1 MB
- Dataset
- Contains an interesting dataset
- Easy to import
- Few or no functions
Data types:
- Binary data,
.Rdataor.rdadata/folder- A single object with the same name as the data file
- Function data
R/sysdata.rda- Data that your functions need
- Raw data,
.xlsx,.csvetcinst/extdatafolder
# Create data in package automatically
usethis::use_data(object, package)
# Manually
save(object, file="path/to/package/data/object.Rdata")
# Access raw data
system.file("extdata","filename.csv", package="packagename")tests/
- What to test?
- That the return value is what is expected given a certain input
- Why test?
- Improved development stability
- Working in group/open source
- Working on big package
- How to test?
- Uses
testthatpackage for testing andusethispackage for setup - Initialize with
use_testthat() - Create test for a function with
use_test("call_me") - Testfile:
tests/testthat/test-call_me.R - Run test with
devtools::test()orcheck()
tests/testthat/test-call_me.R:
test_that("multiplication works", {
expect_equal(2 * 2, 4)
})src/
- Compiled code
RcpprJava
- Scripts
inst/- Dependencies
usethis::use_rcpp()- Edit DESCRIPTION
#' @useDynLib packagename#' @importFrom Rcpp sourceCpp
.cppfile insrc/
src/filename.cpp:
#include <Rcpp.h>
using namespace Rcpp;
// This is a simple example of exporting a C++ function to R. You can
// source this function into an R session using the Rcpp::sourceCpp
// function (or via the Source button on the editor toolbar). Learn
// more about Rcpp at:
//
// http://www.rcpp.org/
// http://adv-r.had.co.nz/Rcpp.html
// http://gallery.rcpp.org/
//
// [[Rcpp::export]]
NumericVector timesTwo(NumericVector x) {
return x * 2;
}src/
- Compiled code
RcpprJava
- Scripts
inst/- Dependencies
usethis::use_rcpp()- Edit DESCRIPTION
#' @useDynLib packagename#' @importFrom Rcpp sourceCpp
.cppfile insrc/pkgbuild::compile_dll()devtools::document()Build & Reload- Add documentation to
.cpp
R/RcppExports.R:
# Generated by using Rcpp::compileAttributes() -> do not edit by hand
# Generator token: 10BE3573-1514-4C36-9D1C-5A225CD40393
timesTwo <- function(x) {
.Call('_typicalr_timesTwo', PACKAGE = 'typicalr', x)
}CRAN and R CMD check
- Comprehensive R Archive Network
- R package repository
- Sign of quality
R CMD check- More than 50 individual checks
- Three messages:
- ERROR: Always fix.
- WARNING: Should probably fix. Definitely for CRAN submit.
- NOTE: Try to solve to CRAN submit, else do not bother.
devtools::check()
Rstudio and Github
git- Version control
- Working in groups
- Rstudio integration
- GitHub
- Unoffical repository
devtools::install_github()- R Package development environment
- Issues

Github Actions
- What it can do
- Integrated with your GitHub repository
- Automates
R CMD check - Test on multiple operating systems
- How it works
- Add a file with instructions to
.github/workflows/workflow-name.yaml - Triggered by action, for example
push - Most common R related workflows available in github r-libs repository
- Add a file with instructions to
Summary
- What is an R package?
- Possible package states
- Package structure:
- Code |
r/ - Metadata |
DESCRIPTION - Documentation |
man/ - Vignettes
- Import & Export |
NAMESPACE - Data |
data/ - Tests |
tests/ - Compiled code |
src/
- Code |
- CRAN and
R CMD check - Rstudio and Github
Thank you! Questions?
_
platform x86_64-pc-linux-gnu
os linux-gnu
major 4
minor 3.2 2024 • SciLifeLab • NBIS • RaukR