Tidy work in Tidyverse
RaukR 2023 • Advanced R for Bioinformatics
27-Jun-2023
Learning Outcomes
When this module is complete, you will:
know what
tidyverseis and a bit about its historybe able to use different pipes, including advanced ones and placeholders
know whether the data you work with are tidy
will be able to load, debug and tidy your data
understand how to combine data sets using
join_*be aware of useful packages within
tidyverse
Tidyverse — what is it all about?
- tidyverse is a collection of packages 📦,
- created by Hadley Wickham,
- has became a de facto standard in data analyses,
- a philosophy of programming or a programming paradigm: everything is about 🌊 the flow of 🧹 tidy data.



?(Tidyverse OR !Tidyverse)
Warning
☠️ There are still some people out there talking about the tidyverse curse though… ☠️
Navigating the balance between base R and the tidyverse is a challenge to learn.
- Robert A. Muenchen
Typical Tidyverse Workflow
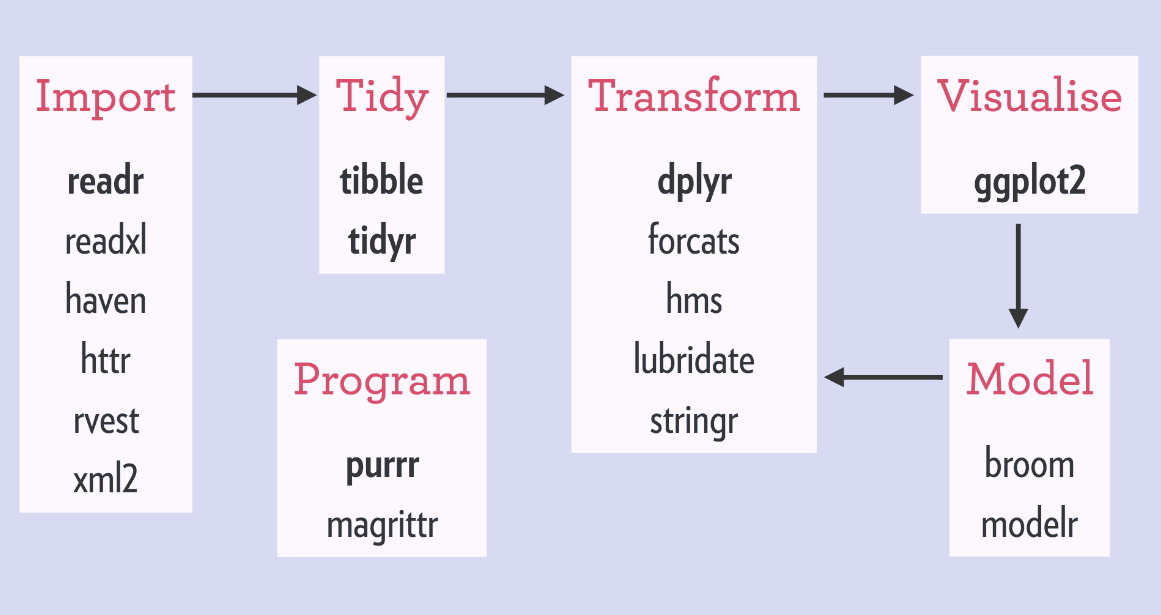
Source: http://www.storybench.org/getting-started-with-tidyverse-in-r/
Introduction to Pipes or Let My Data Flow 🌊
magrittrpackage —tidyverseand beyond- the
%>%pipe
x %>% f\(\equiv\)f(x)x %>% f(y)\(\equiv\)f(x, y)x %>% f %>% g %>% h\(\equiv\)h(g(f(x)))
Introduction to Pipes
Instead of writing this:
write this:
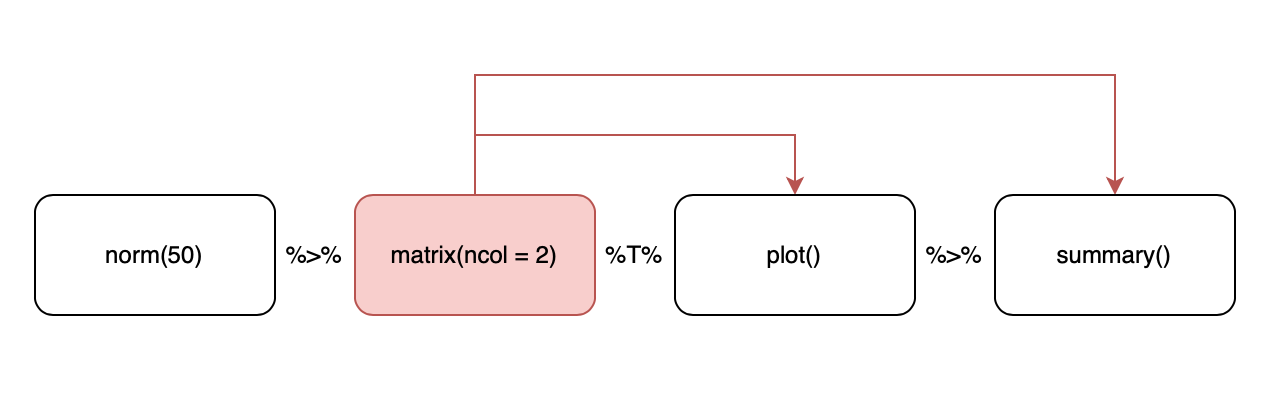
Other Types of Pipes — %T>%
- Provided by
magritter, not intidyverse - When you call a function for its side effects
Other Types of Pipes — %T>%
Other Types of magrittr Pipes — %$%
Error in pmatch(use, c("all.obs", "complete.obs", "pairwise.complete.obs", : object 'Sepal.Width' not foundWe need the %$% pipe with exposition of variables:
This is because cor function does not have the x (data) argument – the very first argument of a pipe-friendly function.
Other Types of magrittr Pipes — %<>%
It exists but can lead to somewhat confusing code! 💀
x %<>% f \(\equiv\) x <- f(x)
Native R pipe
From R >= 4.1.0 we have a native | > pipe that is a bit faster than %>% but currently has no placeholders mechanism.
Placeholders in magrittr Pipes
Sometimes we want to pass the resulting data to other than the first argument of the next function in chain. magritter provides placeholder mechanism for this:
x %>% f(y, .)\(\equiv\)f(y, x),x %>% f(y, z = .)\(\equiv\)f(y, z = x).
Placeholders for nested expressions
But for nested expressions:
x %>% f(a = p(.), b = q(.))\(\equiv\)f(x, a = p(x), b = q(x))
x %>% {f(a = p(.), b = q(.))}\(\equiv\)f(a = p(x), b = q(x))
Placeholders – unary functions
We can even use placeholders as the first element of a pipe:
Functional sequence with the following components:
1. sin(.)
2. cos(.)
Use 'functions' to extract the individual functions. and, indeed the f function works:
Time to do Lab 1.1

Tibbles
tibbleis one of the unifying features of tidyverse,- it is a better
data.framerealization, - objects
data.framecan be coerced totibbleusingas_tibble()
Convert data.frame to tibble
# A tibble: 150 × 5
Sepal.Length Sepal.Width Petal.Length Petal.Width Species
<dbl> <dbl> <dbl> <dbl> <fct>
1 5.1 3.5 1.4 0.2 setosa
2 4.9 3 1.4 0.2 setosa
3 4.7 3.2 1.3 0.2 setosa
4 4.6 3.1 1.5 0.2 setosa
5 5 3.6 1.4 0.2 setosa
6 5.4 3.9 1.7 0.4 setosa
7 4.6 3.4 1.4 0.3 setosa
8 5 3.4 1.5 0.2 setosa
9 4.4 2.9 1.4 0.2 setosa
10 4.9 3.1 1.5 0.1 setosa
# ℹ 140 more rowsTibbles from scratch with tibble
# A tibble: 50 × 4
x y z outcome
<dbl> <dbl> <dbl> <dbl>
1 1 0.152 1.02 0.230
2 1 0.112 1.01 -1.39
3 1 0.283 1.08 0.340
4 1 0.879 1.77 -0.411
5 1 0.182 1.03 -0.731
6 1 0.960 1.92 -0.141
7 1 0.936 1.88 1.67
8 1 0.822 1.68 0.785
9 1 0.745 1.55 -0.360
10 1 0.751 1.56 0.289
# ℹ 40 more rowsMore on Tibbles
- When you print a
tibble:- all columns that fit the screen are shown,
- only the first 10 rows are shown,
- data type for each column is shown.
Tibble printing options
my_tibble %>% print(n = 50, width = Inf),options(tibble.print_min = 15, tibble.print_max = 25),options(dplyr.print_min = Inf),options(tibble.width = Inf)
Subsetting Tibbles
vehicles will be our tibble version of cars
We can access data like this:
Or, alternatively, using placeholders:
Note! Not all old R functions work with tibbles, than you have to use as.data.frame(my_tibble).
Partial Matching
Non-existing Columns
Time to do Lab 1.2

Loading Data
In tidyverse you import data using readr package that provides a number of useful data import functions:
read_delim()a generic function for reading x-delimited files. There are a number of convenience wrappers:read_csv()used to read comma-delimited files,read_csv2()reads semicolon-delimited files,read_tsv()that reads tab-delimited files.
read_fwffor reading fixed-width files with its wrappers:- fwf_widths() for width-based reading,
- fwf_positions() for positions-based reading and
- read_table() for reading white space-delimited fixed-width files.
read_log()for reading Apache-style logs.
Loading Data
The most commonly used read_csv() has some familiar arguments like:
skip– to specify the number of rows to skip (headers),col_names– to supply a vector of column names,comment– to specify what character designates a comment,na– to specify how missing values are represented.
Under the Hood – parse_* Functions
Under the hood, data-reading functions use parse_* functions:
[1] 272555850Parsing Strings
- Strings can be represented in different encodings:
Parsing Factors
- R is using factors to represent categorical variables.
- Supply known levels to
parse_factorso that it warns you when an unknown level is present in the data:
landscapes <- c('mountains', 'swamps', 'seaside')
parse_factor(c('mountains', 'plains', 'seaside', 'swamps'),
levels = landscapes)[1] mountains <NA> seaside swamps
attr(,"problems")
# A tibble: 1 × 4
row col expected actual
<int> <int> <chr> <chr>
1 2 NA value in level set plains
Levels: mountains swamps seasideOther Parsing Functions
parse_
vector,time,number,logical,integer,double,character,date,datetime,guess
Writing to a File
The readr package also provides functions useful for writing tibbled data into a file:
write_csv()write_tsv()write_excel_csv()
They always save:
- Text in UTF-8,
- Dates in ISO8601
But saving in csv (or tsv) does mean you loose information about the type of data in particular columns. You can avoid this by using:
write_rds()andread_rds()to read/write objects in R binary rds format,- Tse
write_feather()andread_feather()from packagefeatherto read/write objects in a fast binary format that other programming languages can access.
Time to do Lab 1.3

Basic Data Transformations with dplyr
Let us create a tibble:
# A tibble: 5 × 10
carat cut color clarity depth table price x y z
<dbl> <ord> <ord> <ord> <dbl> <dbl> <int> <dbl> <dbl> <dbl>
1 0.23 Ideal E SI2 61.5 55 326 3.95 3.98 2.43
2 0.21 Premium E SI1 59.8 61 326 3.89 3.84 2.31
3 0.23 Good E VS1 56.9 65 327 4.05 4.07 2.31
4 0.29 Premium I VS2 62.4 58 334 4.2 4.23 2.63
5 0.31 Good J SI2 63.3 58 335 4.34 4.35 2.75
Picking Observations using filter()
Floating point and tidyverse
Caution
⛵ Be careful with floating point comparisons!
🏴☠️ Also, rows with comparison resulting in NA are skipped by default!
Rearranging Observations using arrange()
# A tibble: 6 × 10
carat cut color clarity depth table price x y z
<dbl> <ord> <ord> <ord> <dbl> <dbl> <int> <dbl> <dbl> <dbl>
1 0.23 Good E VS1 56.9 65 327 4.05 4.07 2.31
2 0.31 Good J SI2 63.3 58 335 4.34 4.35 2.75
3 0.24 Very Good J VVS2 62.8 57 336 3.94 3.96 2.48
4 0.21 Premium E SI1 59.8 61 326 3.89 3.84 2.31
5 0.29 Premium I VS2 62.4 58 334 4.2 4.23 2.63
6 0.23 Ideal E SI2 61.5 55 326 3.95 3.98 2.43Caution
The NAs always end up at the end of the rearranged tibble.
Selecting Variables with select()
Renaming variables
Note
rename is a variant of select, here used with everything() to move x to the beginning and rename it to var_x
# A tibble: 2 × 10
carat cut color clarity depth table price var_x y z
<dbl> <ord> <ord> <ord> <dbl> <dbl> <int> <dbl> <dbl> <dbl>
1 0.23 Ideal E SI2 61.5 55 326 3.95 3.98 2.43
2 0.21 Premium E SI1 59.8 61 326 3.89 3.84 2.31Bring columns to front
Tip
use everything() to bring some columns to the front
# A tibble: 2 × 10
x y z carat cut color clarity depth table price
<dbl> <dbl> <dbl> <dbl> <ord> <ord> <ord> <dbl> <dbl> <int>
1 3.95 3.98 2.43 0.23 Ideal E SI2 61.5 55 326
2 3.89 3.84 2.31 0.21 Premium E SI1 59.8 61 326Create/alter new Variables with mutate
# A tibble: 5 × 9
carat cut color clarity x y z p q
<dbl> <ord> <ord> <ord> <dbl> <dbl> <dbl> <dbl> <dbl>
1 0.23 Ideal E SI2 3.95 3.98 2.43 6.38 10.4
2 0.21 Premium E SI1 3.89 3.84 2.31 6.2 10.0
3 0.23 Good E VS1 4.05 4.07 2.31 6.36 10.4
4 0.29 Premium I VS2 4.2 4.23 2.63 6.83 11.1
5 0.31 Good J SI2 4.34 4.35 2.75 7.09 11.4Create/alter new Variables with transmute 🧙♂️
Caution
Only the transformed variables will be retained.
Group and Summarize
bijou %>% group_by(cut, color) %>% summarize(max_price = max(price),
mean_price = mean(price),
min_price = min(price)) %>% head(n = 5)# A tibble: 5 × 5
# Groups: cut [3]
cut color max_price mean_price min_price
<ord> <ord> <int> <dbl> <int>
1 Good E 327 327 327
2 Good J 335 335 335
3 Very Good J 336 336 336
4 Premium E 326 326 326
5 Premium I 334 334 334Other data manipulation tips
# A tibble: 4 × 2
cut count
<ord> <int>
1 Good 2
2 Very Good 1
3 Premium 2
4 Ideal 1When you need to regroup within the same pipe, use ungroup().
The Concept of Tidy Data
- Each and every observation is represented as exactly one row,
- Each and every variable is represented by exactly one column,
- Thus each data table cell contains only one value.

Usually data are untidy in only one way. However, if you are unlucky, they are really untidy and thus a pain to work with…
Tidy Data

Are these data tidy?
| Sepal.Length | Sepal.Width | Petal.Length | Petal.Width | Species |
|---|---|---|---|---|
| 5.1 | 3.5 | 1.4 | 0.2 | setosa |
| 4.9 | 3.0 | 1.4 | 0.2 | setosa |
| 4.7 | 3.2 | 1.3 | 0.2 | setosa |
| Species | variable | value |
|---|---|---|
| setosa | Sepal.Length | 5.1 |
| setosa | Sepal.Length | 4.9 |
| setosa | Sepal.Length | 4.7 |
Tidy Data

Are these data tidy?
| Sepal.L.W | Petal.L.W | Species |
|---|---|---|
| 5.1/3.5 | 1.4/0.2 | setosa |
| 4.9/3 | 1.4/0.2 | setosa |
| 4.7/3.2 | 1.3/0.2 | setosa |
| Sepal.Length | 5.1 | 4.9 | 4.7 | 4.6 |
| Sepal.Width | 3.5 | 3.0 | 3.2 | 3.1 |
| Petal.Length | 1.4 | 1.4 | 1.3 | 1.5 |
| Petal.Width | 0.2 | 0.2 | 0.2 | 0.2 |
| Species | setosa | setosa | setosa | setosa |
Tidying Data with pivot_longer
If some of your column names should be values of a variable, use pivot_longer (old gather):
Tidying Data with pivot_wider
If some of your observations are scattered across many rows, use pivot_wider (old spread):
# A tibble: 9 × 5
cut price clarity dimension measurement
<ord> <int> <ord> <chr> <dbl>
1 Ideal 326 SI2 x 3.95
2 Premium 326 SI1 x 3.89
3 Good 327 VS1 x 4.05
4 Ideal 326 SI2 y 3.98
5 Premium 326 SI1 y 3.84
6 Good 327 VS1 y 4.07
7 Ideal 326 SI2 z 2.43
8 Premium 326 SI1 z 2.31
9 Good 327 VS1 z 2.31Tidying Data with separate
If some of your columns contain more than one value, use separate:
# A tibble: 2 × 4
cut price clarity dim
<ord> <int> <ord> <chr>
1 Ideal 326 SI2 3.95/3.98/2.43
2 Premium 326 SI1 3.89/3.84/2.31# A tibble: 2 × 6
cut price clarity x y z
<ord> <int> <ord> <dbl> <dbl> <dbl>
1 Ideal 326 SI2 3.95 3.98 2.43
2 Premium 326 SI1 3.89 3.84 2.31Note
Here, sep is here interpreted as the position to split on. It can also be a regular expression or a delimiting string/character. Pretty flexible approach!
Tidying Data with unite
If some of your columns contain more than one value
# A tibble: 5 × 7
cut price clarity_prefix clarity_suffix x y z
<ord> <int> <chr> <chr> <dbl> <dbl> <dbl>
1 Ideal 326 SI 2 3.95 3.98 2.43
2 Premium 326 SI 1 3.89 3.84 2.31
3 Good 327 VS 1 4.05 4.07 2.31
4 Premium 334 VS 2 4.2 4.23 2.63
5 Good 335 SI 2 4.34 4.35 2.75Completing Missing Values Using complete
# A tibble: 7 × 4
cut continent clarity price
<ord> <chr> <ord> <int>
1 Fair Eur <NA> NA
2 Good Eur VS1 327
3 Good Eur SI2 335
4 Very Good Eur VVS2 336
5 Premium Eur SI1 326
6 Premium Eur VS2 334
7 Ideal Eur SI2 326Combining Datasets
Often, we need to combine a number of data tables (relational data) to get the full picture of the data. Here different types of joins come to help:
- mutating joins that add new variables to data table
Abased on matching observations (rows) from data tableB
- filtering joins that filter observations from data table
Abased on whether they match observations in data tableB
- set operations that treat observations in
AandBas elements of a set.
Let us create two example tibbles that share a key:
| key | x |
|---|---|
| a | A1 |
| b | A2 |
| c | A3 |
| e | A4 |
| key | y |
|---|---|
| a | B1 |
| b | NA |
| c | B3 |
| d | B4 |
The Joins Family — inner_join
| key | x |
|---|---|
| a | A1 |
| b | A2 |
| c | A3 |
| e | A4 |
| key | y |
|---|---|
| a | B1 |
| b | NA |
| c | B3 |
| d | B4 |
The Joins Family — left_join
| key | x |
|---|---|
| a | A1 |
| b | A2 |
| c | A3 |
| e | A4 |
| key | y |
|---|---|
| a | B1 |
| b | NA |
| c | B3 |
| d | B4 |
The Joins Family — right_join
| key | x |
|---|---|
| a | A1 |
| b | A2 |
| c | A3 |
| e | A4 |
| key | y |
|---|---|
| a | B1 |
| b | NA |
| c | B3 |
| d | B4 |
The Joins Family — full_join
| key | x |
|---|---|
| a | A1 |
| b | A2 |
| c | A3 |
| e | A4 |
| key | y |
|---|---|
| a | B1 |
| b | NA |
| c | B3 |
| d | B4 |
Some Other Friends
stringrfor string manipulation and regular expressionsforcatsfor working with factorslubridatefor working with dates
Thank you! Questions?
_
platform x86_64-pc-linux-gnu
os linux-gnu
major 4
minor 2.3 2023 • SciLifeLab • NBIS • RaukR