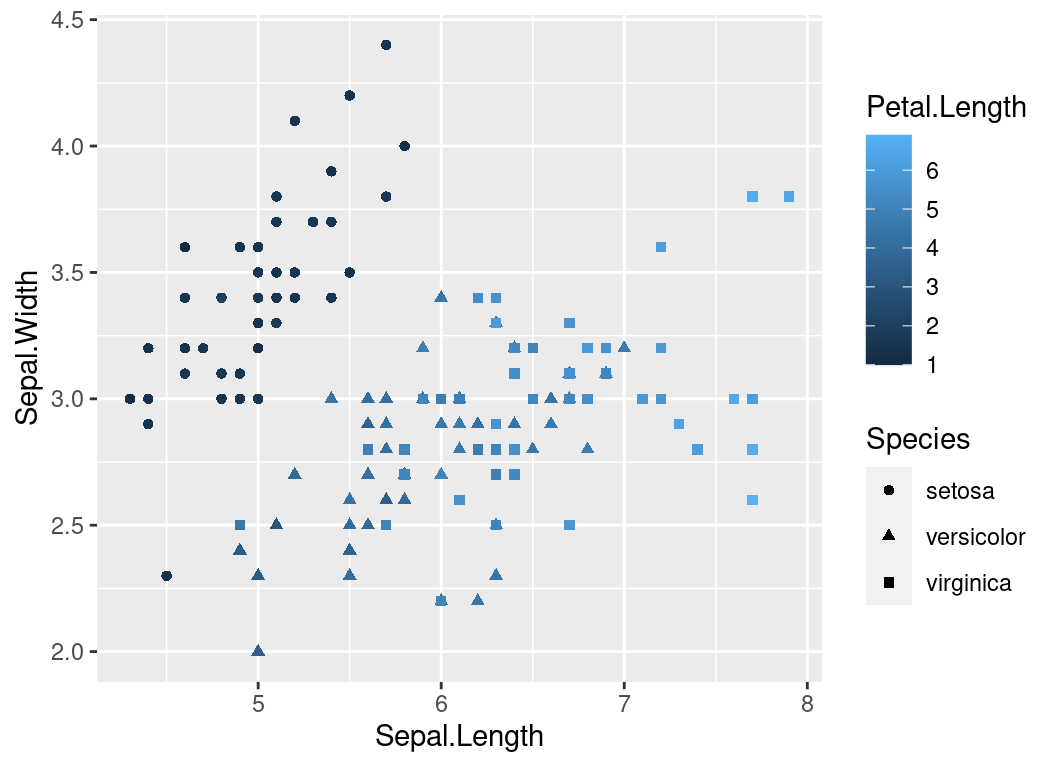
Plotting with ggplot2
RaukR 2023 • Advanced R for Bioinformatics
Roy Francis
27-Jun-2023
Graphs
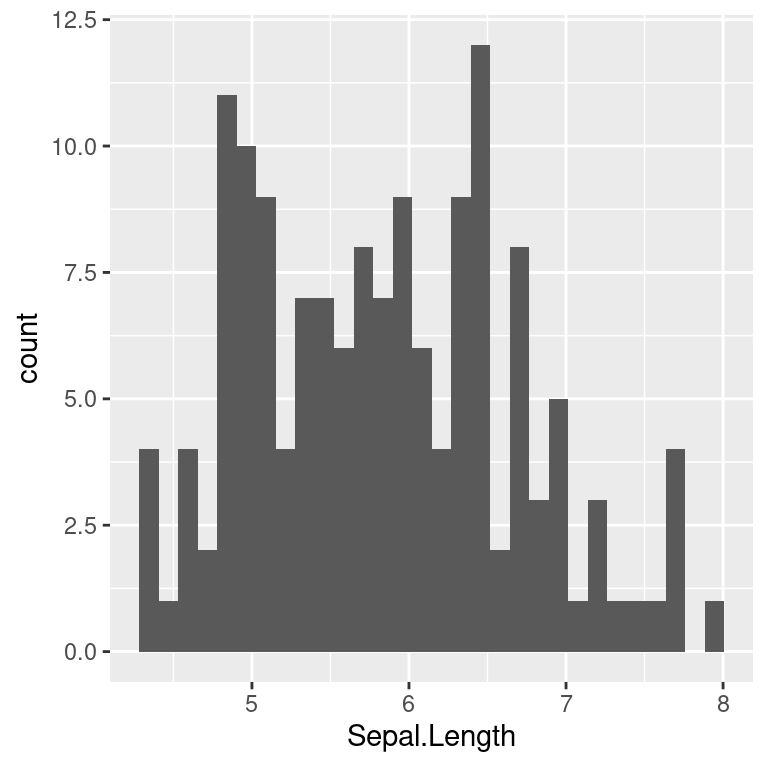
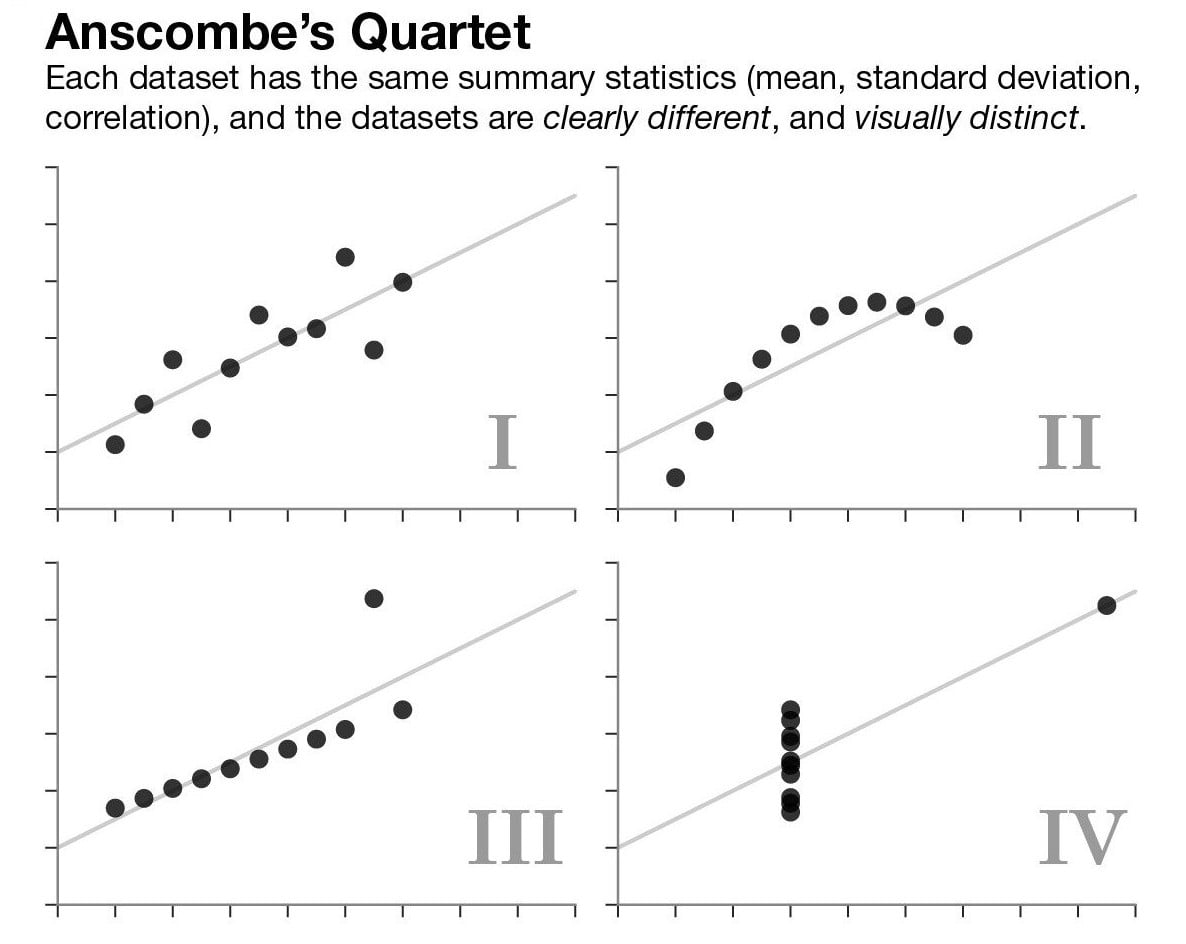
Graphing is an essential part of data analyses. Data with same summary statistics can look very different when plotted out.

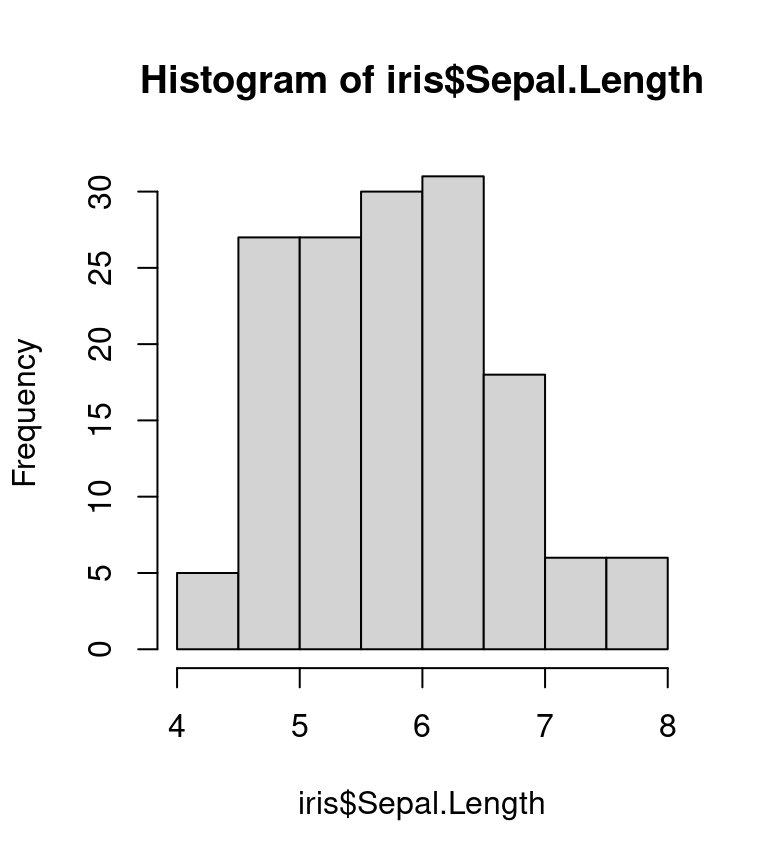
R graphics
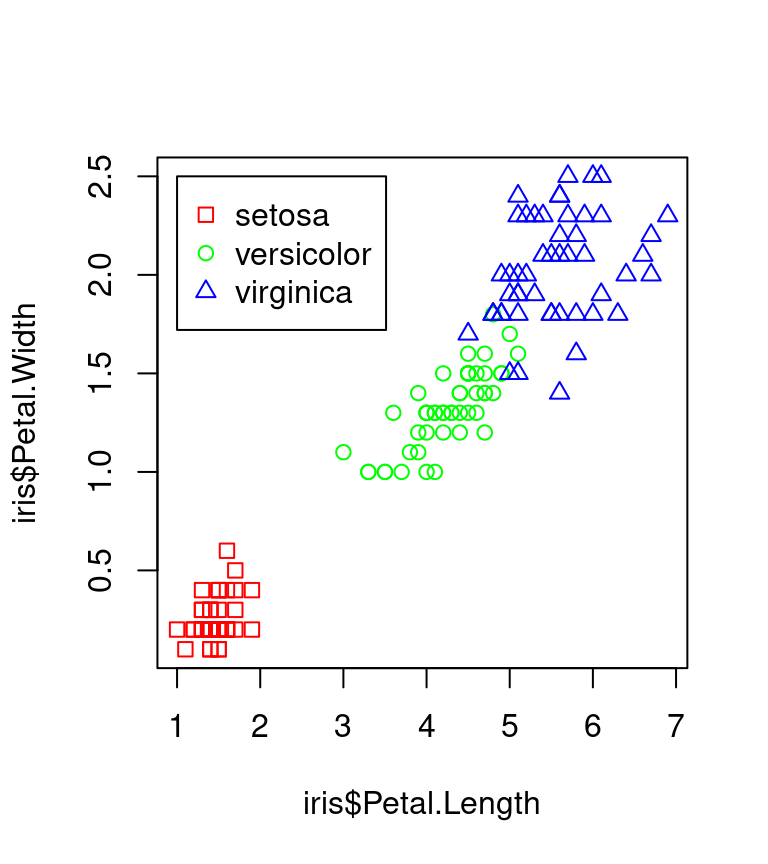
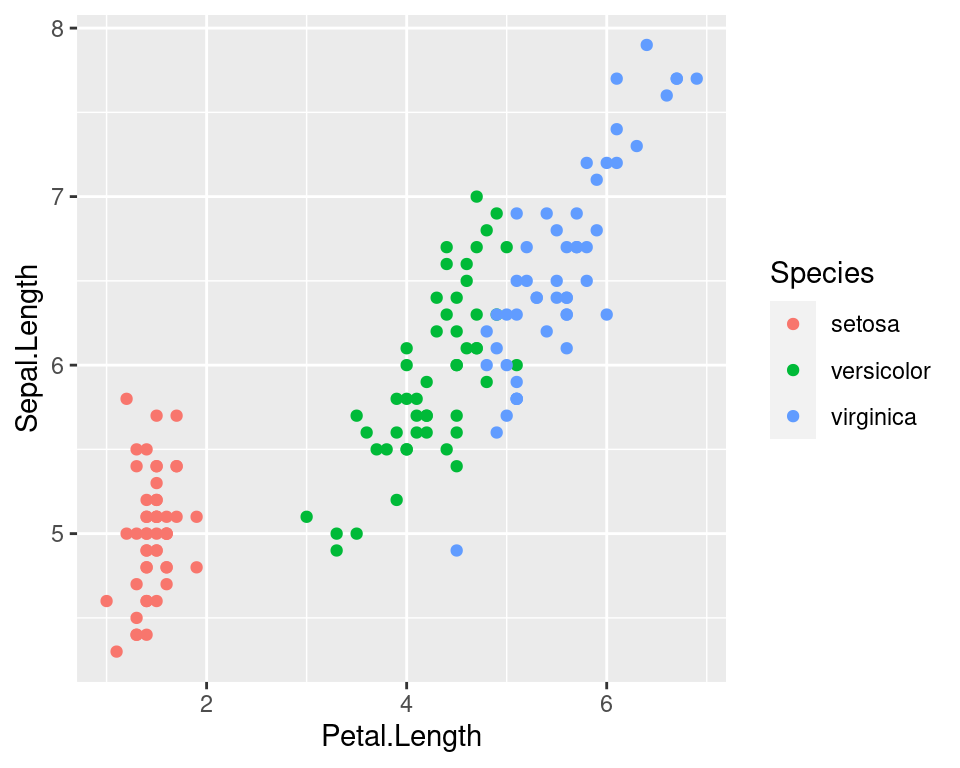
ggplot2 vs Base Graphics
ggplot2 vs Base Graphics
Why ggplot2?
- Consistent code for any type of plot (almost!)
- Flexible and modular (Add/remove components)
- Automatic legends, colors etc
- Save plot objects
- Themes for reusing styles
- Numerous add-ons/extensions
- Nearly complete structured graphing solution
- Adapted to other programming languages
Grammar Of Graphics
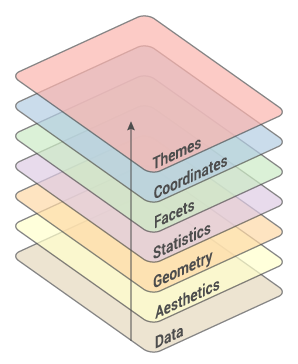

- Created by Hadley Wickham in 2005
- Data: Input data
- Geom: A geometry representing data. Points, Lines etc
- Aesthetic: Visual characteristics of the geometry. Size, Color, Shape etc
- Scale: How visual characteristics are converted to display values
- Statistics: Statistical transformations. Counts, Means etc
- Coordinates: Numeric system to determine position of geometry. Cartesian, Polar etc
- Facets: Split data into subsets
Building A Graph: Syntax

Building A Graph
Building A Graph
Building A Graph
Building A Graph
Data • iris
- Input data is always an R
data.frameobject
| Sepal.Length | Sepal.Width | Petal.Length | Petal.Width | Species |
|---|---|---|---|---|
| 5.1 | 3.5 | 1.4 | 0.2 | setosa |
| 4.9 | 3.0 | 1.4 | 0.2 | setosa |
| 4.7 | 3.2 | 1.3 | 0.2 | setosa |
| 4.6 | 3.1 | 1.5 | 0.2 | setosa |
| 5.0 | 3.6 | 1.4 | 0.2 | setosa |
| 5.4 | 3.9 | 1.7 | 0.4 | setosa |
'data.frame': 150 obs. of 5 variables:
$ Sepal.Length: num 5.1 4.9 4.7 4.6 5 5.4 4.6 5 4.4 4.9 ...
$ Sepal.Width : num 3.5 3 3.2 3.1 3.6 3.9 3.4 3.4 2.9 3.1 ...
$ Petal.Length: num 1.4 1.4 1.3 1.5 1.4 1.7 1.4 1.5 1.4 1.5 ...
$ Petal.Width : num 0.2 0.2 0.2 0.2 0.2 0.4 0.3 0.2 0.2 0.1 ...
$ Species : Factor w/ 3 levels "setosa","versicolor",..: 1 1 1 1 1 1 1 1 1 1 ...Data • diamonds
| carat | cut | color | clarity | depth | table | price | x | y | z |
|---|---|---|---|---|---|---|---|---|---|
| 0.23 | Ideal | E | SI2 | 61.5 | 55 | 326 | 3.95 | 3.98 | 2.43 |
| 0.21 | Premium | E | SI1 | 59.8 | 61 | 326 | 3.89 | 3.84 | 2.31 |
| 0.23 | Good | E | VS1 | 56.9 | 65 | 327 | 4.05 | 4.07 | 2.31 |
| 0.29 | Premium | I | VS2 | 62.4 | 58 | 334 | 4.20 | 4.23 | 2.63 |
| 0.31 | Good | J | SI2 | 63.3 | 58 | 335 | 4.34 | 4.35 | 2.75 |
| 0.24 | Very Good | J | VVS2 | 62.8 | 57 | 336 | 3.94 | 3.96 | 2.48 |
tibble [53,940 × 10] (S3: tbl_df/tbl/data.frame)
$ carat : num [1:53940] 0.23 0.21 0.23 0.29 0.31 0.24 0.24 0.26 0.22 0.23 ...
$ cut : Ord.factor w/ 5 levels "Fair"<"Good"<..: 5 4 2 4 2 3 3 3 1 3 ...
$ color : Ord.factor w/ 7 levels "D"<"E"<"F"<"G"<..: 2 2 2 6 7 7 6 5 2 5 ...
$ clarity: Ord.factor w/ 8 levels "I1"<"SI2"<"SI1"<..: 2 3 5 4 2 6 7 3 4 5 ...
$ depth : num [1:53940] 61.5 59.8 56.9 62.4 63.3 62.8 62.3 61.9 65.1 59.4 ...
$ table : num [1:53940] 55 61 65 58 58 57 57 55 61 61 ...
$ price : int [1:53940] 326 326 327 334 335 336 336 337 337 338 ...
$ x : num [1:53940] 3.95 3.89 4.05 4.2 4.34 3.94 3.95 4.07 3.87 4 ...
$ y : num [1:53940] 3.98 3.84 4.07 4.23 4.35 3.96 3.98 4.11 3.78 4.05 ...
$ z : num [1:53940] 2.43 2.31 2.31 2.63 2.75 2.48 2.47 2.53 2.49 2.39 ...Data • Format

Wide
| Sepal.Length | Sepal.Width | Petal.Length | Petal.Width | Species |
|---|---|---|---|---|
| 5.1 | 3.5 | 1.4 | 0.2 | setosa |
| 4.9 | 3.0 | 1.4 | 0.2 | setosa |
| 4.7 | 3.2 | 1.3 | 0.2 | setosa |
Long
| Species | variable | value |
|---|---|---|
| setosa | Sepal.Length | 5.1 |
| setosa | Sepal.Length | 4.9 |
| setosa | Sepal.Length | 4.7 |
Geoms
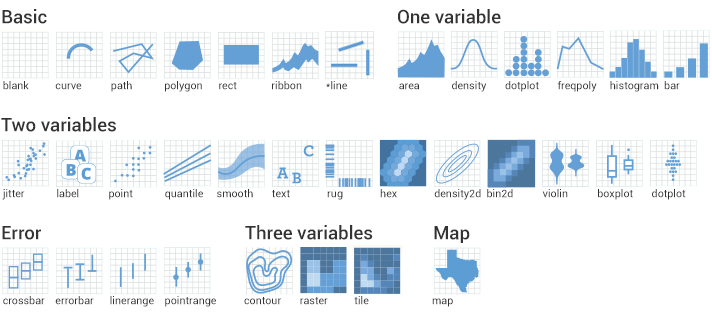
geoms
Stats
- Stats compute new variables from input data.
Stats
- Plots can be built with stats.

Stats
- Stats have default geoms.
| plot | stat | geom |
|---|---|---|
| histogram | bin | bar |
| smooth | smooth | line |
| boxplot | boxplot | boxplot |
| density | density | line |
| freqpoly | freqpoly | line |
Use args(geom_bar) to check arguments.
Aesthetics
Aesthetics
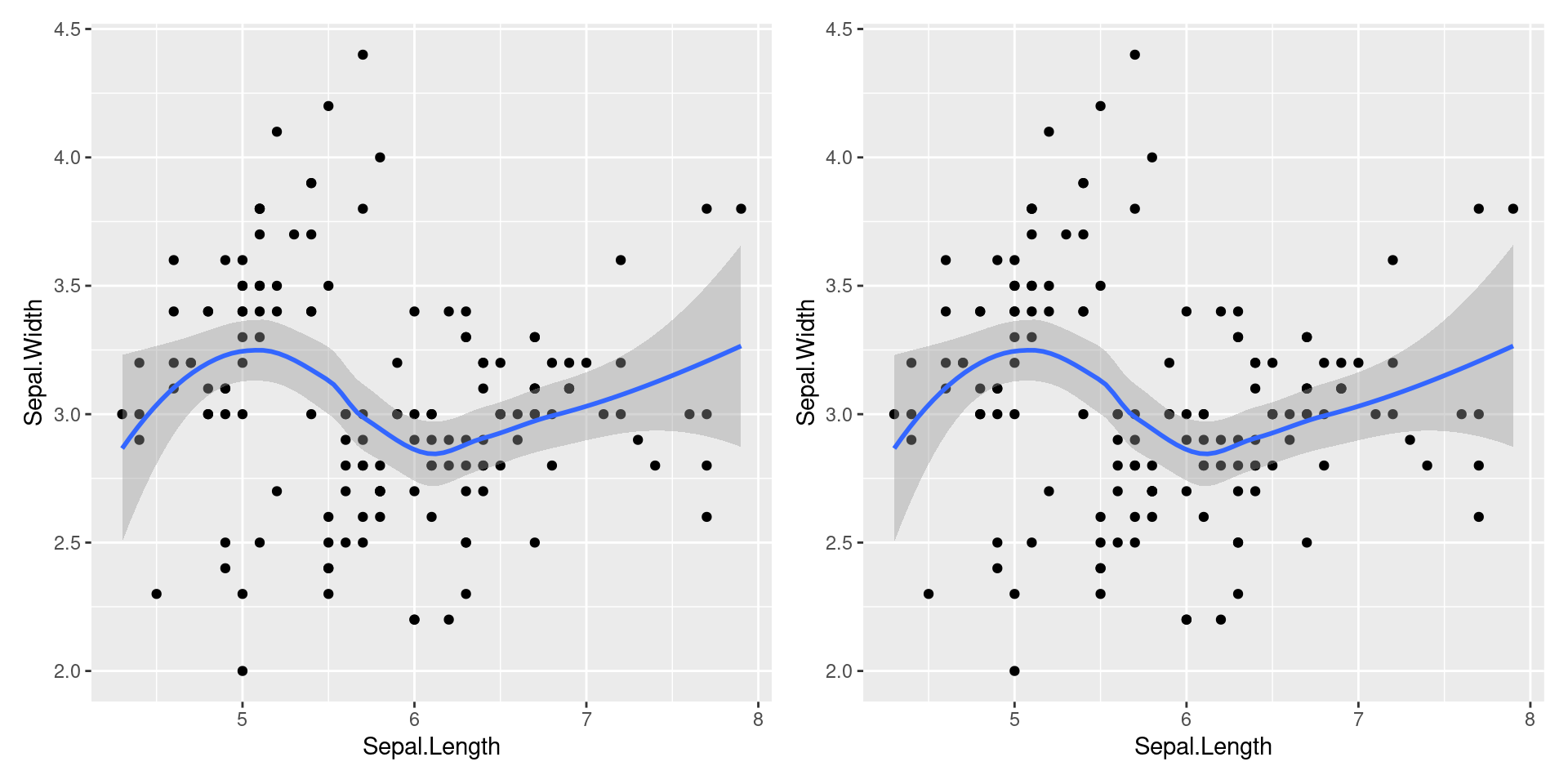
Multiple Geoms
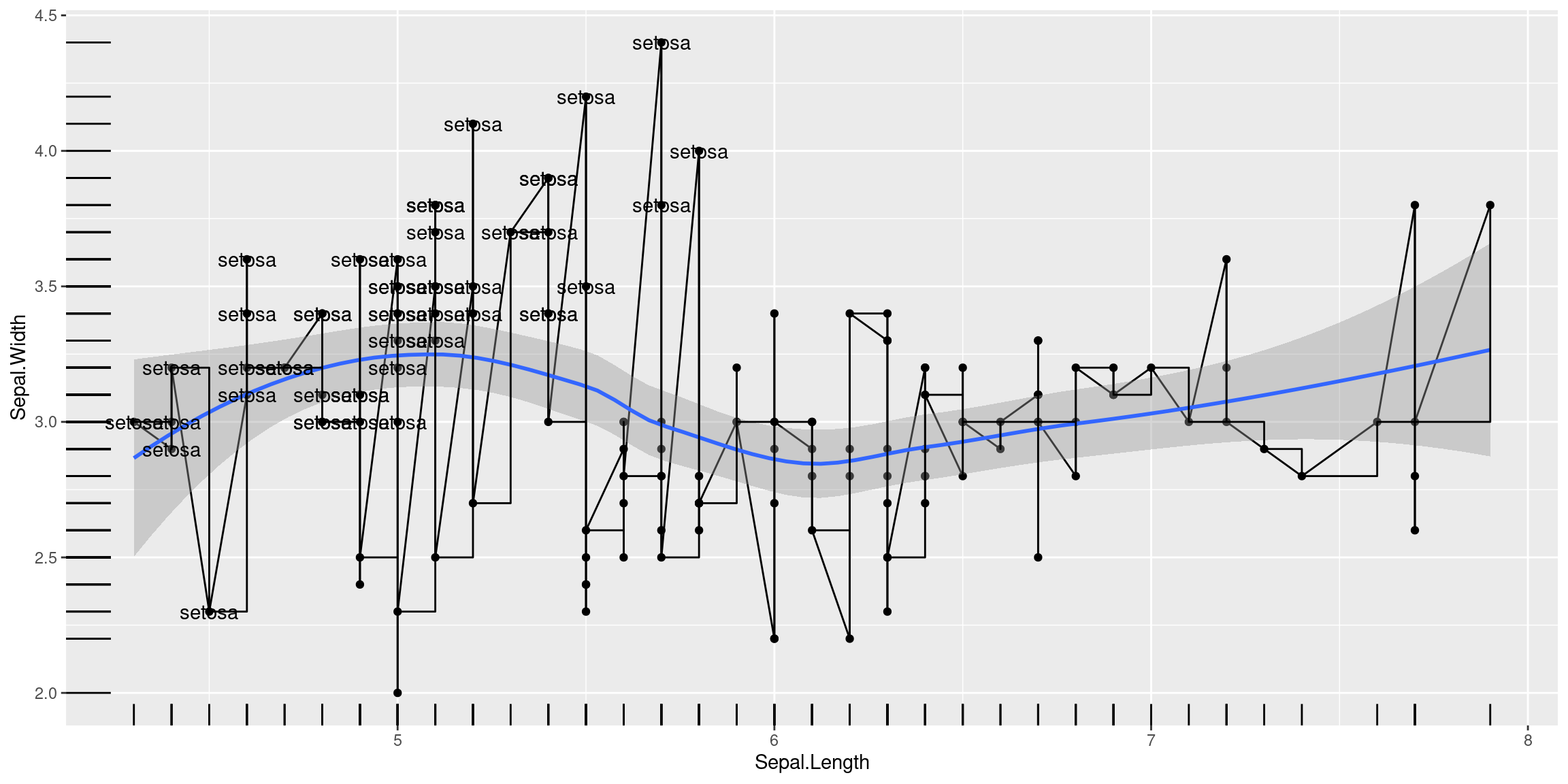
Just because you can doesn’t mean you should!
Scales • Discrete Colors
- scales: position, color, fill, size, shape, alpha, linetype
- syntax:
scale_<aesthetic>_<type>
Scales • Continuous Colors
- In RStudio, type
scale_, then press TAB
Scales • Shape
Scales • Axes
- scales: x, y
- syntax:
scale_<axis>_<type> - arguments: name, limits, breaks, labels
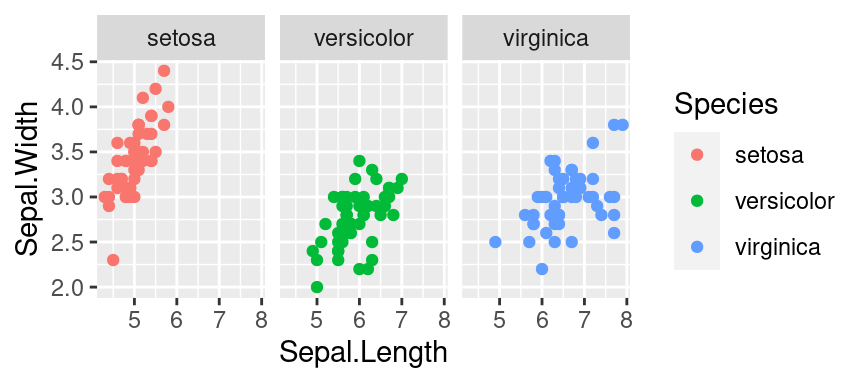
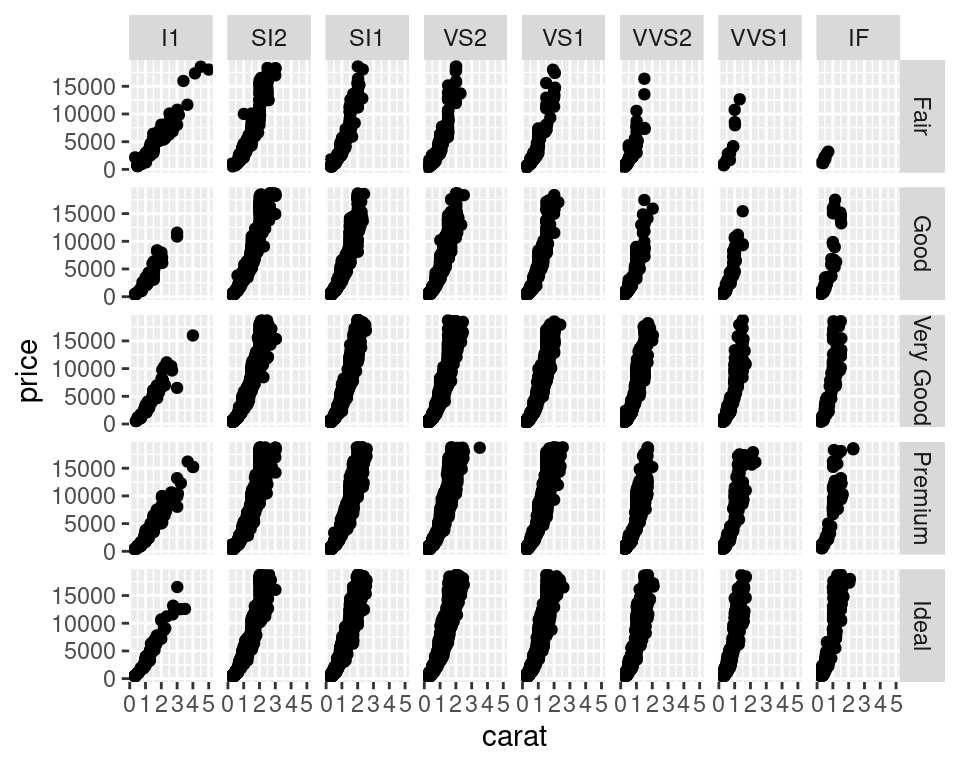
Facets • facet_wrap
- Split to subplots based on variable(s), Faceting in one dimension
Facets • facet_grid
Coordinate Systems
Theming
- Modify non-data plot elements/appearance
- Axis labels, panel colors, legend appearance etc
Theme • Legend
Theme • Text
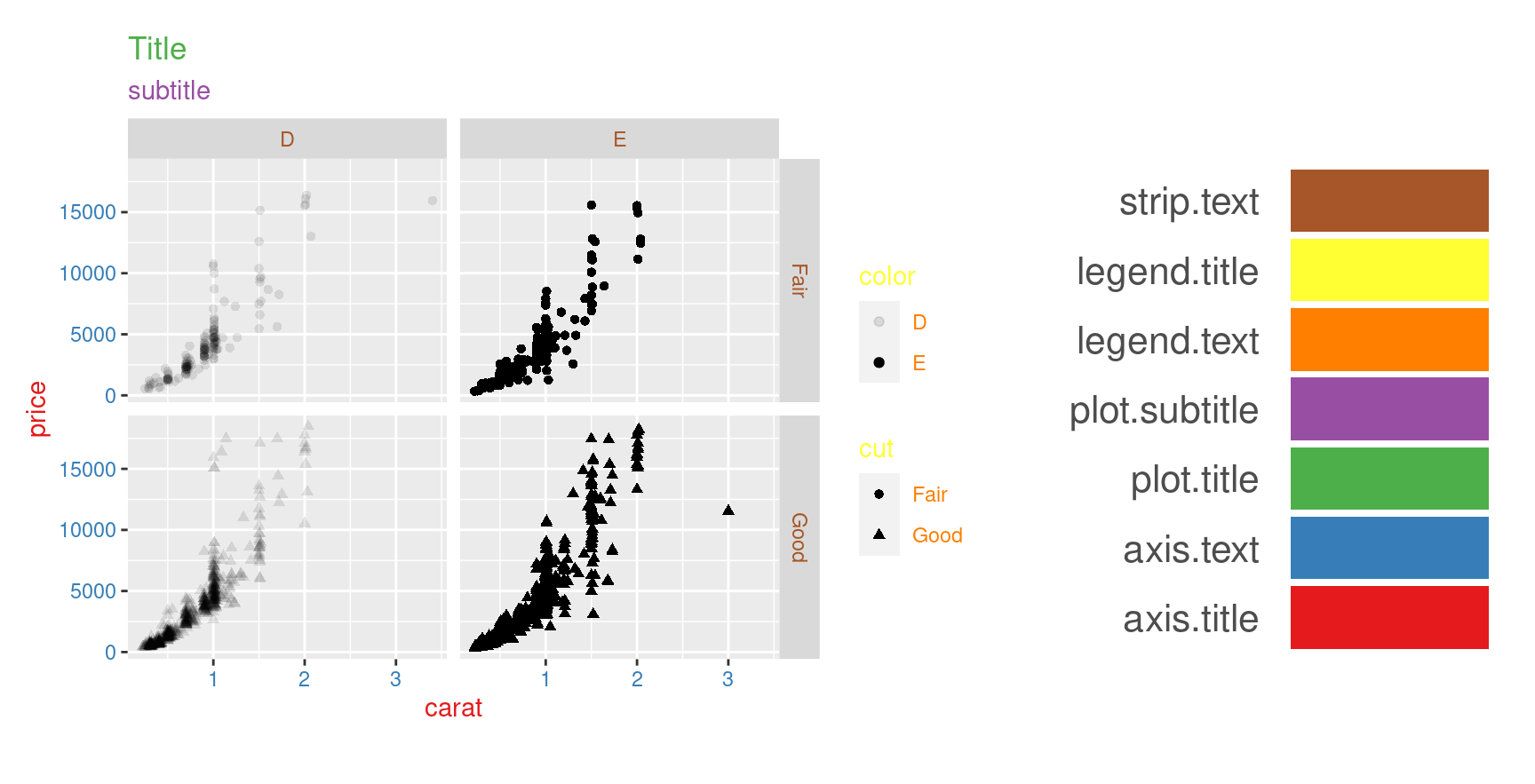
p <- p + theme(
axis.title=element_text(color="#e41a1c"),
axis.text=element_text(color="#377eb8"),
plot.title=element_text(color="#4daf4a"),
plot.subtitle=element_text(color="#984ea3"),
legend.text=element_text(color="#ff7f00"),
legend.title=element_text(color="#ffff33"),
strip.text=element_text(color="#a65628")
)
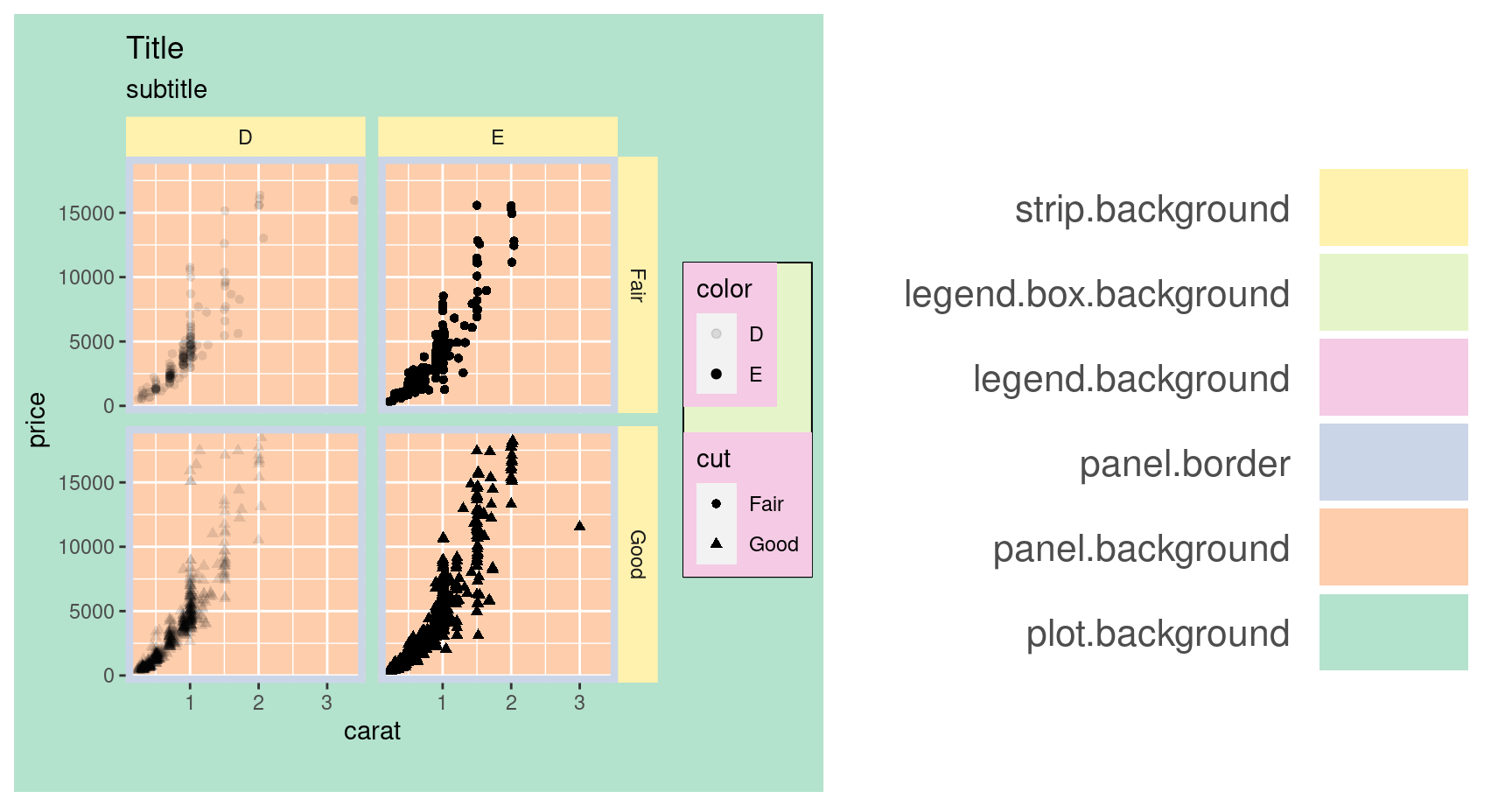
Theme • Rect
p <- p + theme(
plot.background=element_rect(fill="#b3e2cd"),
panel.background=element_rect(fill="#fdcdac"),
panel.border=element_rect(fill=NA,color="#cbd5e8",size=3),
legend.background=element_rect(fill="#f4cae4"),
legend.box.background=element_rect(fill="#e6f5c9"),
strip.background=element_rect(fill="#fff2ae")
)
Theme • Reuse
newtheme <- theme_bw() + theme(
axis.ticks=element_blank(), panel.background=element_rect(fill="white"),
panel.grid.minor=element_blank(), panel.grid.major.x=element_blank(),
panel.grid.major.y=element_line(size=0.3,color="grey90"), panel.border=element_blank(),
legend.position="top", legend.justification="right"
)Professional themes
Position
Saving plots
ggplot2plots can be saved just like base plots
ggplot2package offers a convenient function
- Note that default units in
pngis pixels while inggsaveit’s inches
Combining Plots

Refer to patchwork documentation.
Interactive
- Convert
ggplot2object to interactive HTML
Extensions
- patchwork: Combining plots
- ggrepel: Text labels including overlap control
- ggforce: Circles, splines, hulls, voronoi etc
- ggpmisc: Miscellaneaous features
- ggthemes: Set of extra themes
- ggthemr: More themes
- ggsci: Color palettes for scales
- ggmap: Dedicated to mapping
- ggraph: Network graphs
- ggiraph: Converting ggplot2 to interactive graphics
A collection of ggplot extension packages: https://exts.ggplot2.tidyverse.org/.
Curated list of ggplot2 links: https://github.com/erikgahner/awesome-ggplot2.
Help
Thank you! Questions?
_
platform x86_64-pc-linux-gnu
os linux-gnu
major 4
minor 2.3 2023 • SciLifeLab • NBIS • RaukR