Overview
Teaching: 45 min
Exercises: 30 minQuestions
How can I create simple plots with ggplot?
What is faceting in ggplot?
How can I change the aestetics (e.g. axis labels and color) of a plot?
Objectives
Produce scatter plots, boxplots, and barplots using ggplot.
Set universal plot settings.
Describe what faceting is and apply faceting in ggplot.
Modify the aesthetics of an existing ggplot plot (including axis labels and color).
Build complex and customized plots from data in a data frame.
Getting ready for plotting
We start by loading the required packages. ggplot2 is included in
the tidyverse package.
library(tidyverse)
If not still in the workspace, load the Hawks dataset used in the previous episode.
hawks <- read_csv("data_raw/Hawks.csv")
Plotting with ggplot2
ggplot2 is a plotting package that makes it simple to create complex plots
from data in a data frame. It provides a more programmatic interface for
specifying what variables to plot, how they are displayed, and general visual
properties. Therefore, we only need minimal changes if the underlying data
change or if we decide to change from a bar plot to a scatterplot. This helps in
creating publication quality plots with minimal amounts of adjustments and
tweaking.
ggplot2 plots work best with data in the ‘long’ format, i.e. a column for
every dimension, and a row for every observation. Well-structured data will save
you lots of time when making figures with ggplot2.
ggplot graphics are built layer by layer by adding new elements. Adding layers in this fashion allows for extensive flexibility and customization of plots.
To build a ggplot, we will use the following basic template that can be used for different types of plots:
ggplot(data = <DATA>, mapping = aes(<MAPPINGS>)) + <GEOM_FUNCTION>()
- use the
ggplot()function and bind the plot to a specific data frame using thedataargument:
ggplot(data = Hawks)
- define an aesthetic mapping (using the aesthetic (
aes) function), by selecting the variables to be plotted and specifying how to present them in the graph, e.g. as x/y positions, or characteristics such as size, shape, color, etc:
ggplot(data = hawks, mapping = aes(x = Weight, y = Weight))
-
add ‘geoms’ (geometric objects) – graphical representations of the data in the plot (points, lines, bars).
ggplot2offers many different geoms; we will use some common ones today, including:geom_point()for scatter plots, dot plots, etc.geom_boxplot()for, well, boxplots!geom_bar()for displaying relations between numeric and categorical variables.
To add a geom to the plot use + operator. Let’s first try geom_point():
ggplot(data = hawks, mapping = aes(x = Weight, y = Wing)) +
geom_point()
Warning: Removed 11 rows containing missing values (geom_point).
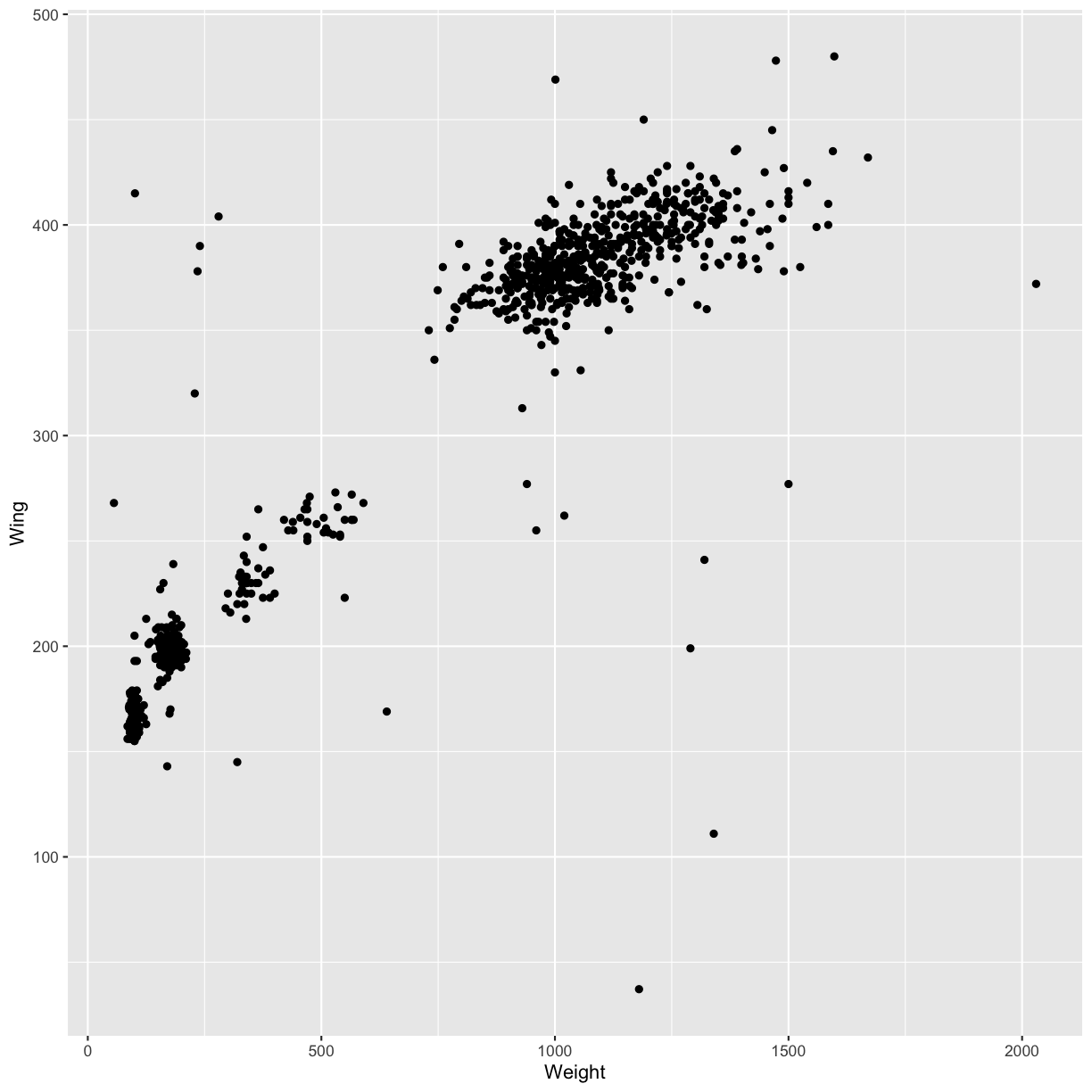
The + in the ggplot2 package is particularly useful because it allows
you to modify existing ggplot objects. This means you can easily set up plot
“templates” and conveniently explore different types of plots, so the above
plot can also be generated with code like this:
# Assign plot to a variable
weight_vs_wing_plot <- ggplot(data = hawks, mapping = aes(x = Weight, y = Wing))
# Draw the plot
weight_vs_wing_plot +
geom_point()
Notes
- Anything you put in the
ggplot()function can be seen by any geom layers that you add (i.e. these are universal plot settings). This includes the x- and y-axis you set up inaes(). - You can also specify aesthetics for a given geom independently of the
aesthetics defined globally in the
ggplot()function. - The
+sign used to add layers must be placed at the end of each line containing a layer. If, instead, the+sign is added in the line before the other layer,ggplot2will not add the new layer and will return an error message.
# This is the correct syntax for adding layers
weight_vs_wing_plot +
geom_point()
# This will not add the new layer and will return an error message
weight_vs_wing_plot
+ geom_point()
Challenge 4.1
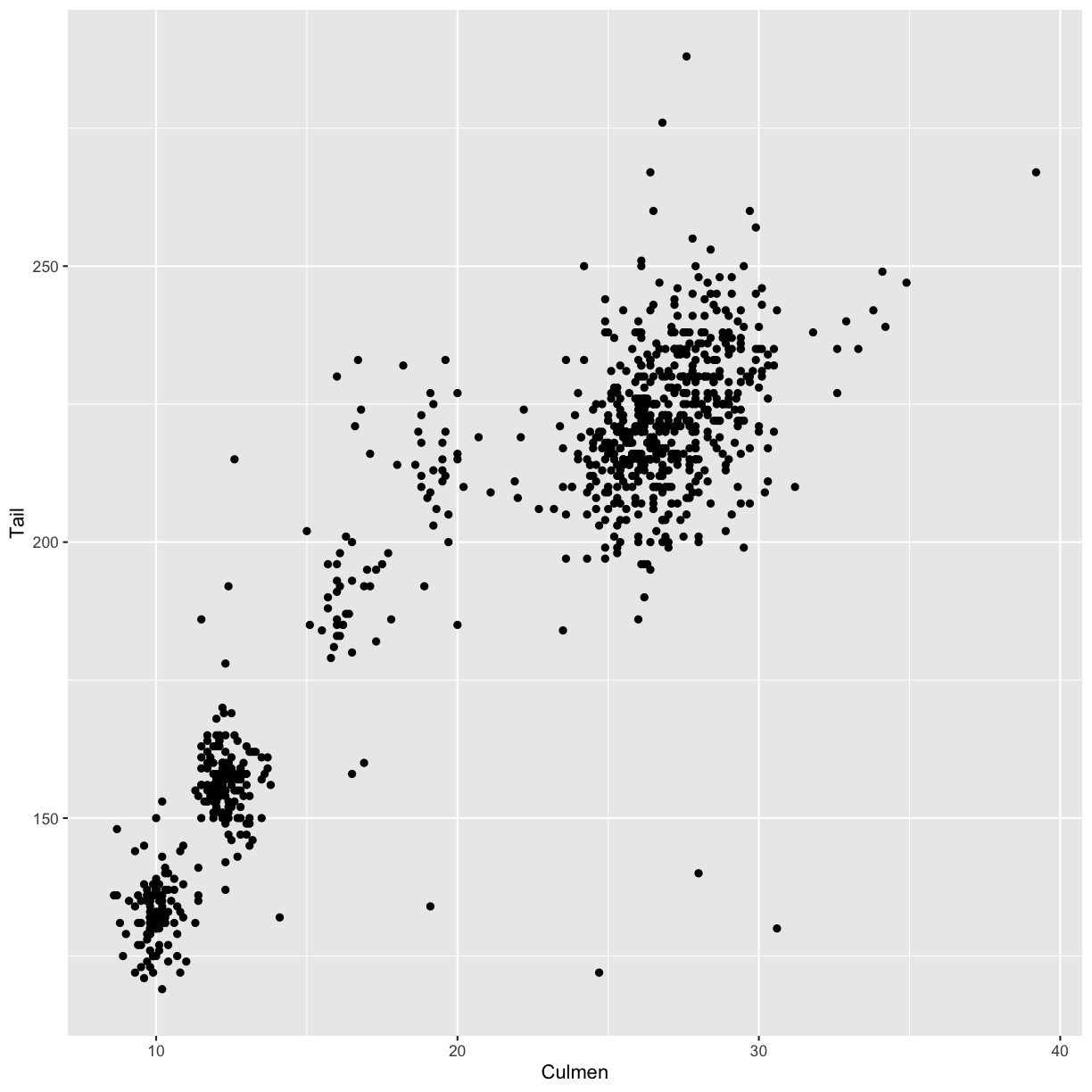
Create a scatter plot with the culmen length (column
Culmen) plotted against the tail length (columnTail).Solution
ggplot(data = hawks, mapping = aes(x = Culmen, y = Tail)) + geom_point()Warning: Removed 7 rows containing missing values (geom_point).
Building your plots iteratively
Building plots with ggplot2 is typically an iterative process. We start by
defining the dataset we’ll use, lay out the axes, and choose a geom:
ggplot(data = hawks, mapping = aes(x = Weight, y = Wing)) +
geom_point()
Warning: Removed 11 rows containing missing values (geom_point).
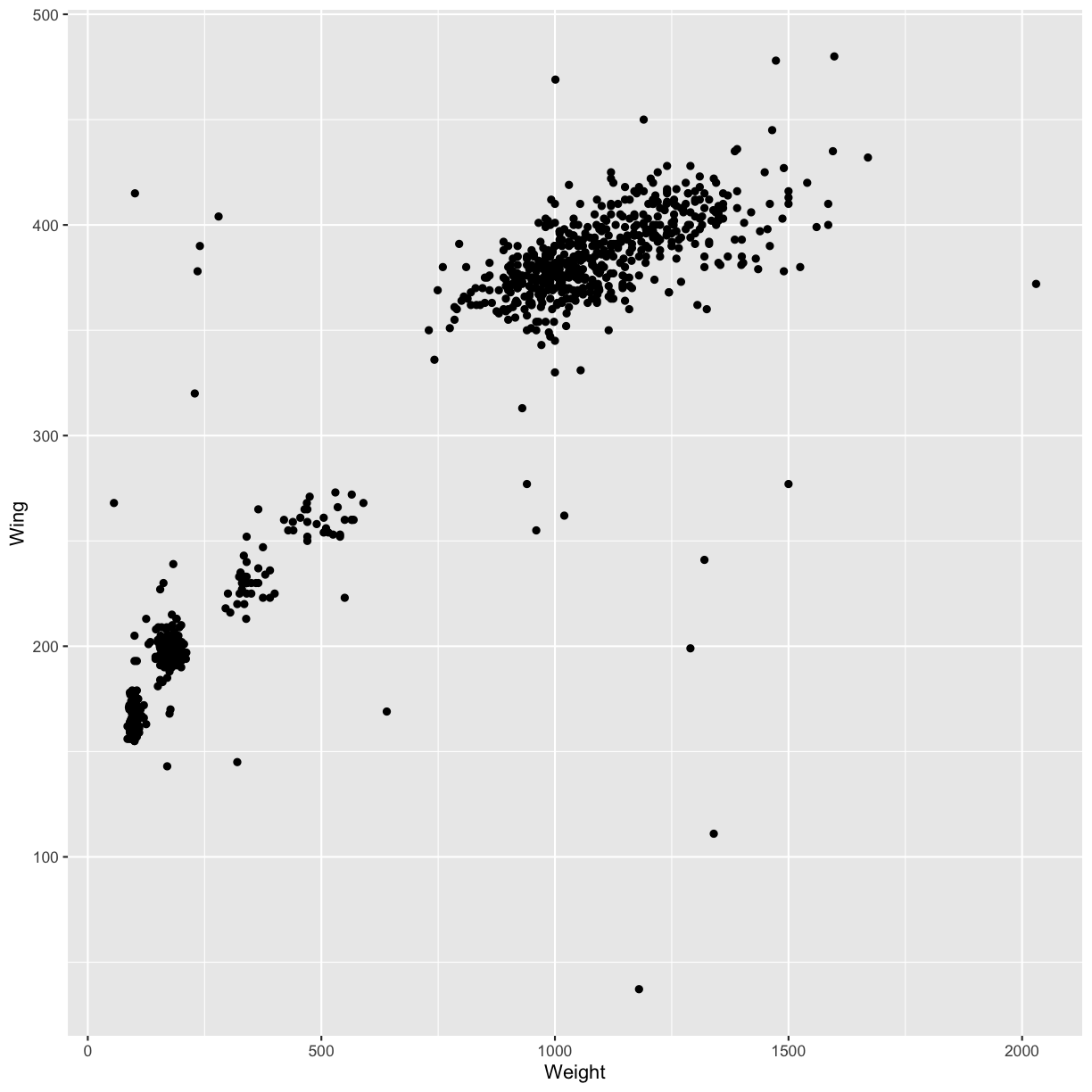
Then, we start modifying this plot to extract more information from it. For
instance, we can add transparency (alpha) to avoid overplotting:
ggplot(data = hawks, mapping = aes(x = Weight, y = Wing)) +
geom_point(alpha = 0.5)
Warning: Removed 11 rows containing missing values (geom_point).
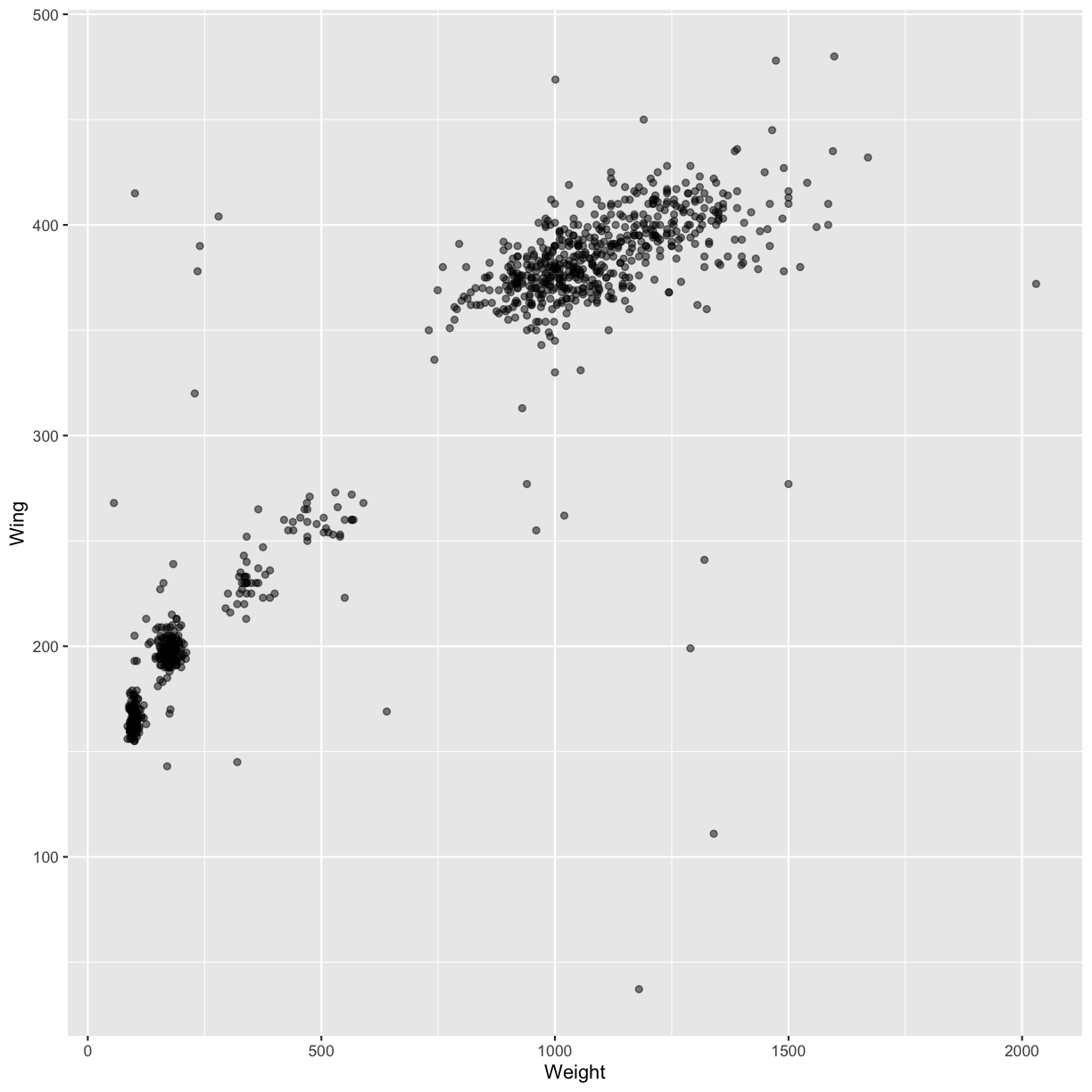
We can also add colors for all the points:
ggplot(data = hawks, mapping = aes(x = Weight, y = Wing)) +
geom_point(alpha = 0.5, color = "blue")
Warning: Removed 11 rows containing missing values (geom_point).
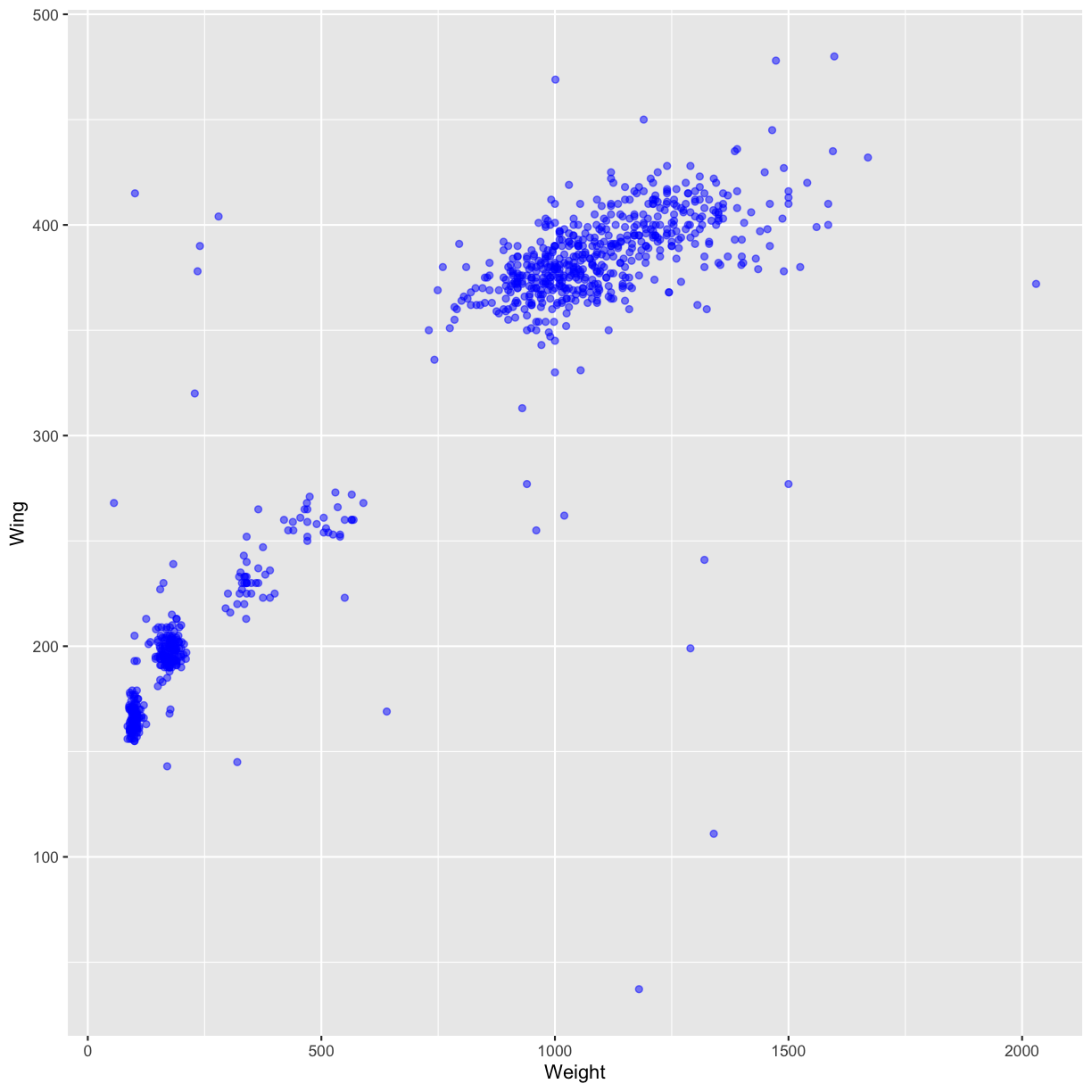
Or to color each point in the plot differently, you could use a vector as an
input to the argument color. ggplot2 will provide a different color
corresponding to different values in the vector. Here is an example where we
color with Species:
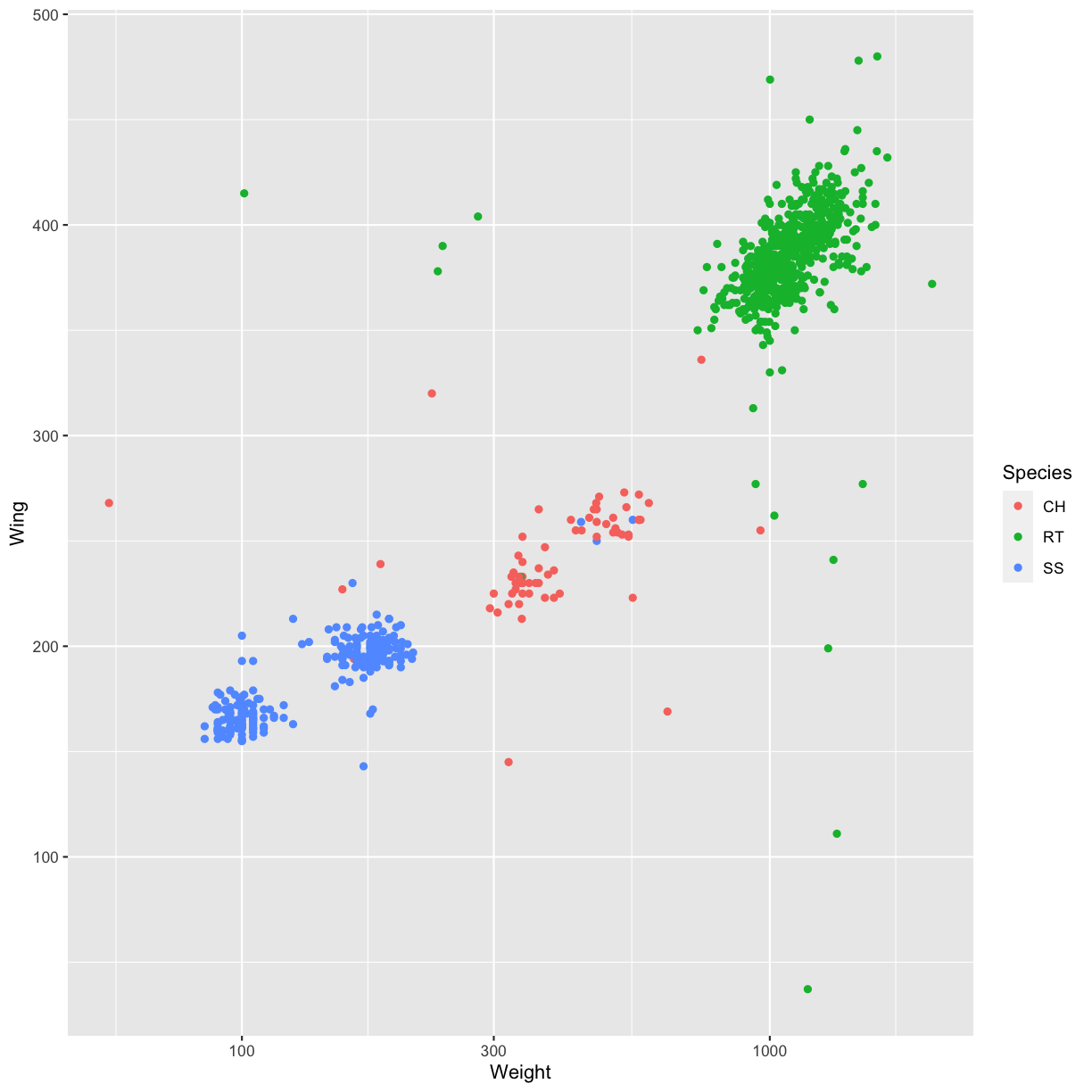
ggplot(data = hawks, mapping = aes(x = Weight, y = Wing)) +
geom_point(alpha = 0.5, aes(color = Species))
Warning: Removed 11 rows containing missing values (geom_point).
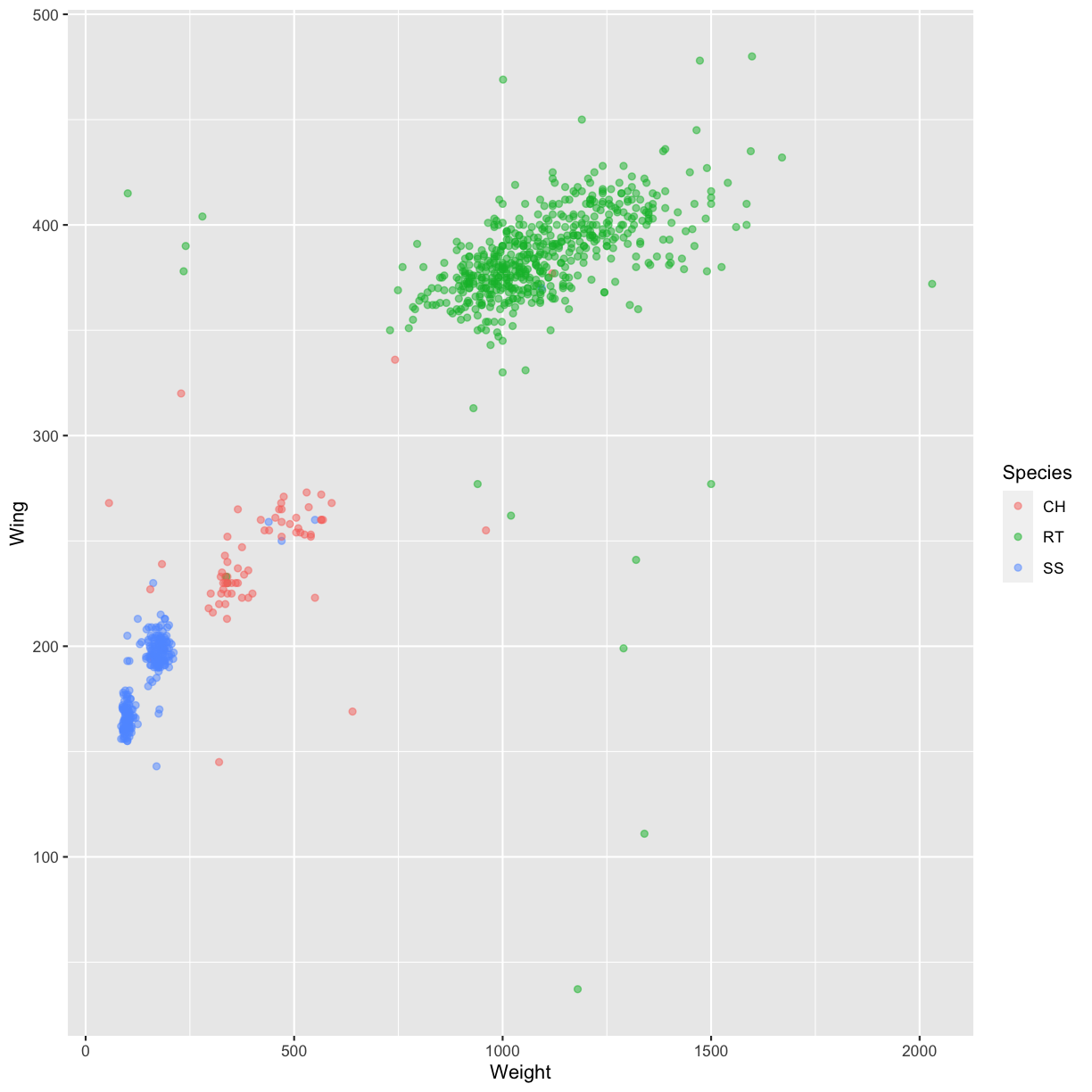
Challenge 4.2
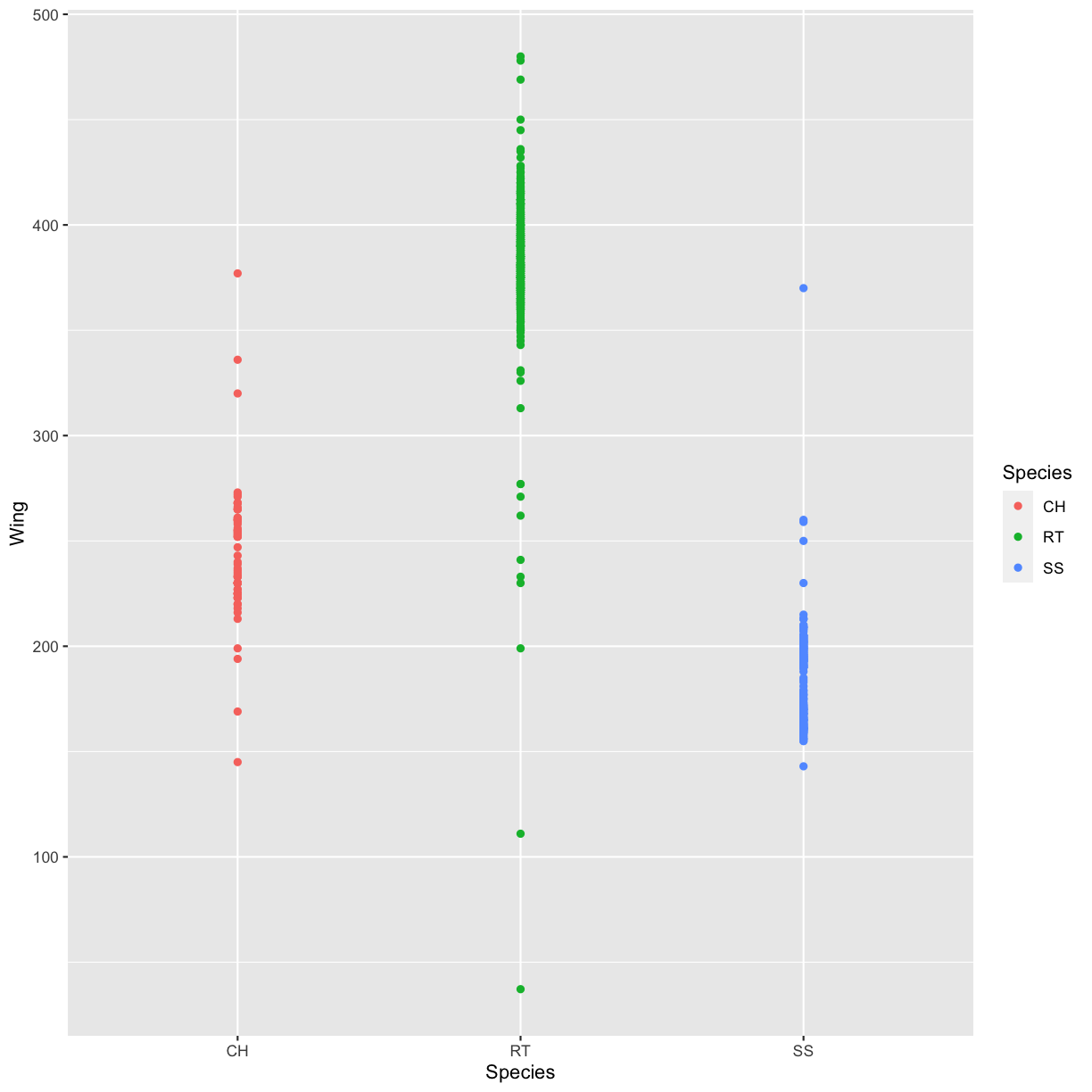
Use what you just learned to create a scatter plot of the species (column
Species) against the wing length (columnWing). Is this a good way to show this type of data?Solution
ggplot(data = hawks, mapping = aes(x = Species, y = Wing)) + geom_point(aes(color = Species))Warning: Removed 1 rows containing missing values (geom_point).
Boxplot
Another useful way to visualize and compare distributions across groups is the boxplot. Here we will first create a boxplot that visualizes the distribution of wing lengths within each Species:
ggplot(data = hawks, mapping = aes(x = Species, y = Wing)) +
geom_boxplot()
Warning: Removed 1 rows containing non-finite values (stat_boxplot).
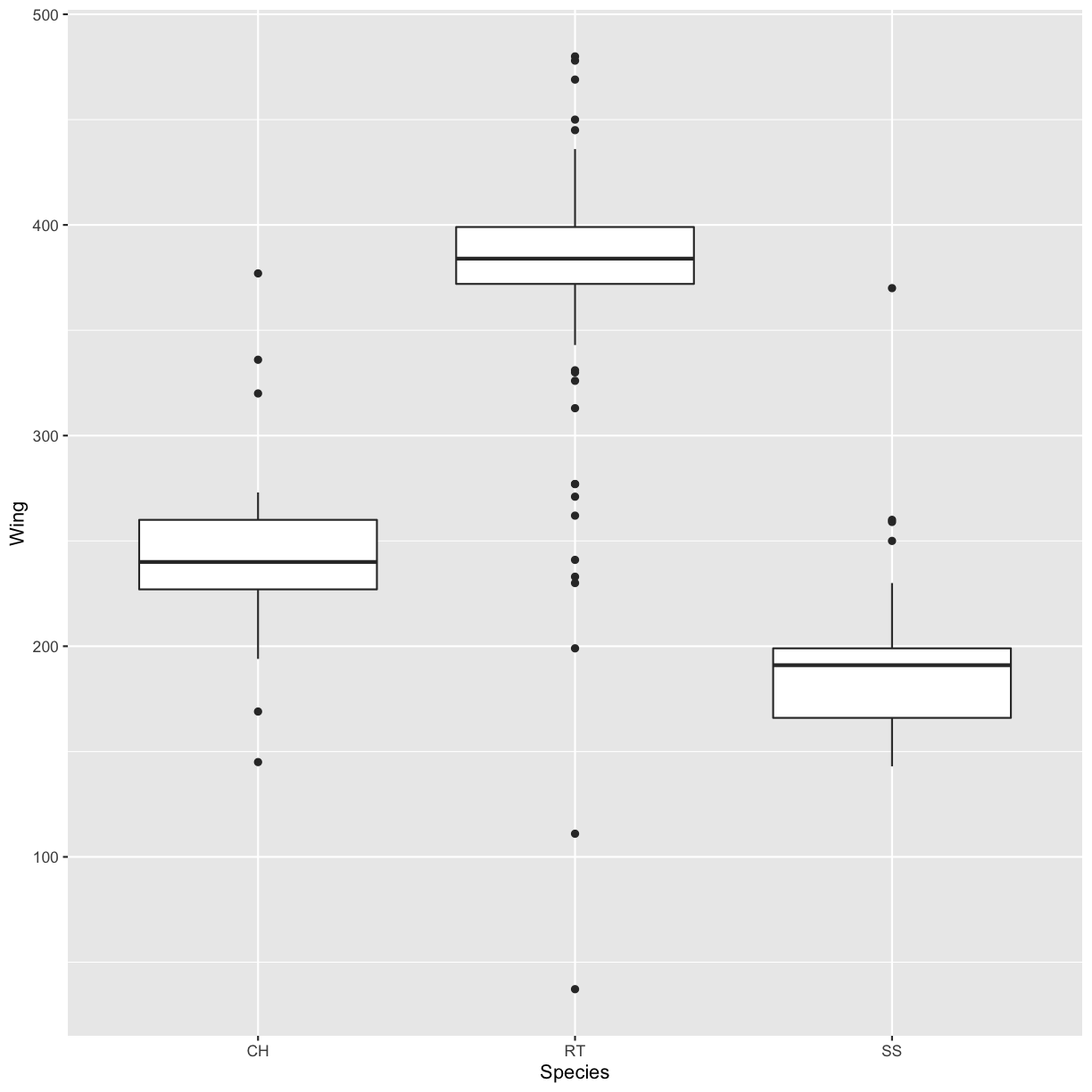
By adding points to the boxplot, we can have a better idea of the number of counts and of their distribution:
ggplot(data = hawks, mapping = aes(x = Species, y = Wing)) +
geom_boxplot(alpha = 0) +
geom_jitter(alpha = 0.5, color = "tomato")
Warning: Removed 1 rows containing non-finite values (stat_boxplot).
Warning: Removed 1 rows containing missing values (geom_point).
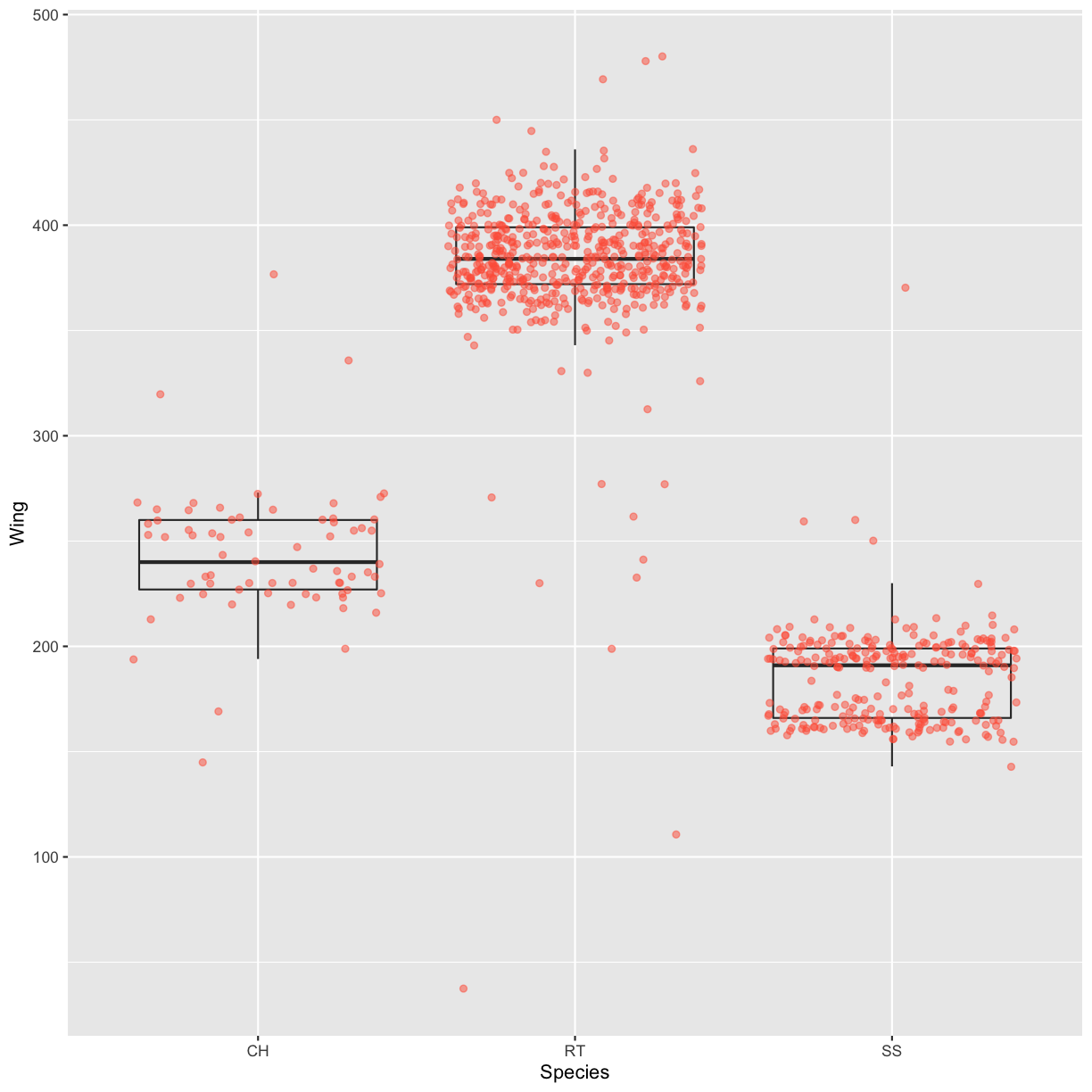
Notice how the boxplot layer is behind the jitter layer? What do you need to change in the code to put the boxplot in front of the points such that it’s not hidden?
Challenges 4.3
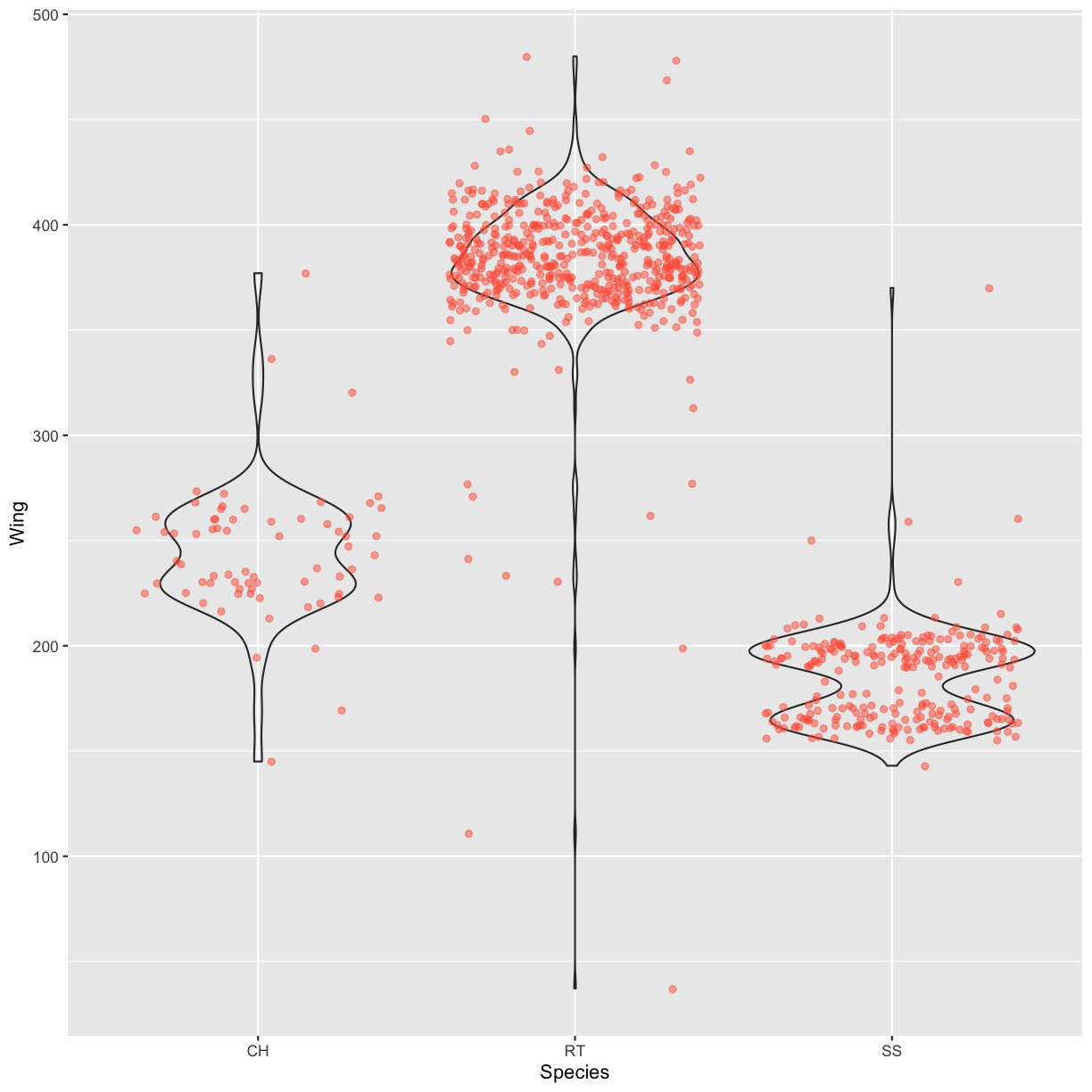
Boxplots are useful summaries, but hide the shape of the distribution. For example, if there is a bimodal distribution, it would not be observed with a boxplot. An alternative to the boxplot is the violin plot (sometimes known as a beanplot), where the shape (of the density of points) is drawn. Replace the box plot with a violin plot; see
geom_violin(). Modify the code below to show a violin plot instead.ggplot(data = hawks, mapping = aes(x = Species, y = Wing)) + geom_boxplot(alpha = 0) + geom_jitter(alpha = 0.5, color = "tomato")Solution
ggplot(data = hawks, mapping = aes(x = Species, y = Wing)) + geom_violin(alpha = 0) + geom_jitter(alpha = 0.5, color = "tomato")Warning: Removed 1 rows containing non-finite values (stat_ydensity).Warning: Removed 1 rows containing missing values (geom_point).
In many types of data, it is important to consider the scale of the observations. For example, it may be worth changing the scale of the axis to better distribute the observations in the space of the plot. Changing the scale of the axes is done similarly to adding/modifying other components.
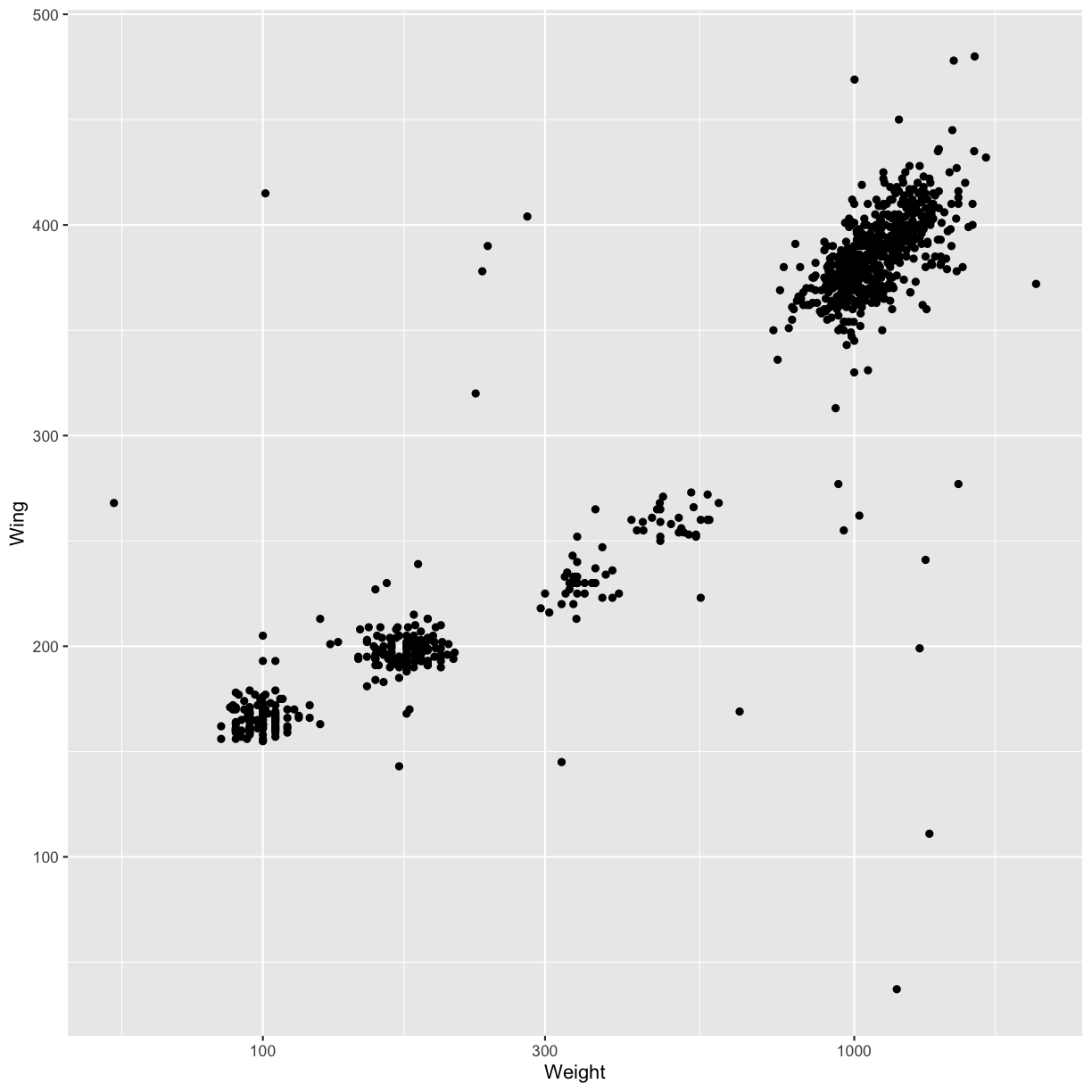
- Modify the code below so that weight is shown on a log 10 scale; see
scale_x_log10().ggplot(data = hawks, mapping = aes(x = Weight, y = Wing)) + geom_point(alpha = 0)Solution
ggplot(data = hawks, mapping = aes(x = Weight, y = Wing)) + geom_point() + scale_x_log10()Warning: Removed 11 rows containing missing values (geom_point).
- Add color to the data points on your plot according to the Species.
Solution
ggplot(data = hawks, mapping = aes(x = Weight, y = Wing)) + geom_point(aes(color = Species)) + scale_x_log10()Warning: Removed 11 rows containing missing values (geom_point).
Barplot
Another common type of plot is the barplot. This kind of plot can be created
with geom_bar(). In order to create a barplot, we will first prepare a
suitable dataset:
sex_counts <- hawks %>% count(Sex)
sex_counts
# A tibble: 3 × 2
Sex n
<chr> <int>
1 F 174
2 M 158
3 <NA> 576
Let’s then create a barplot from the tiny dataset that we just created:
ggplot(sex_counts, aes(x = Sex, y = n)) +
geom_bar(stat = "identity")
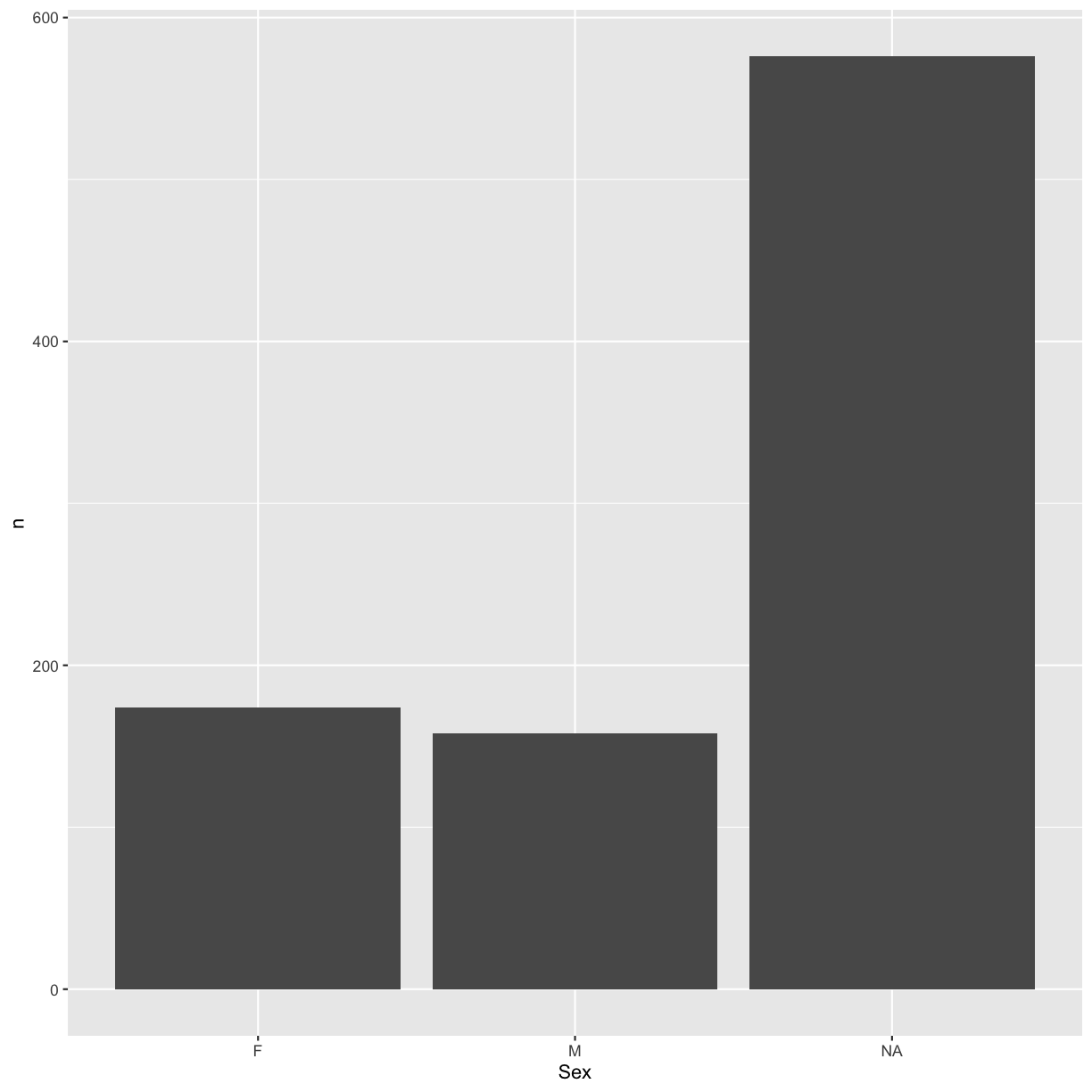
In the code above, we used the argument stat = "identity" instead of the
default value bin. This means that the height of the bar will be represented
by the count in each category.
We can improve the plot by using different fill colors for the sexes:
ggplot(sex_counts, aes(x = Sex, y = n, fill = Sex)) +
geom_bar(stat = "identity")
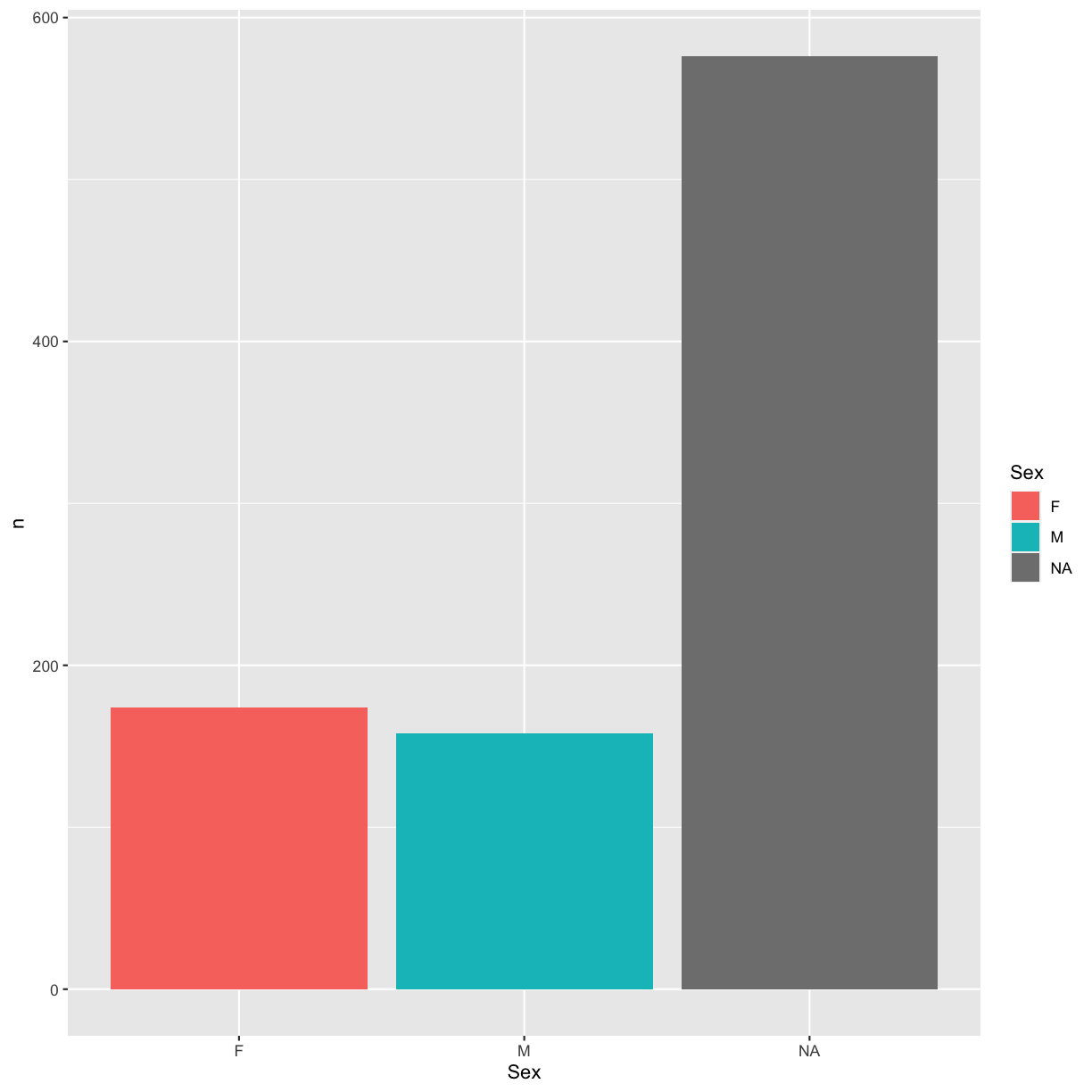
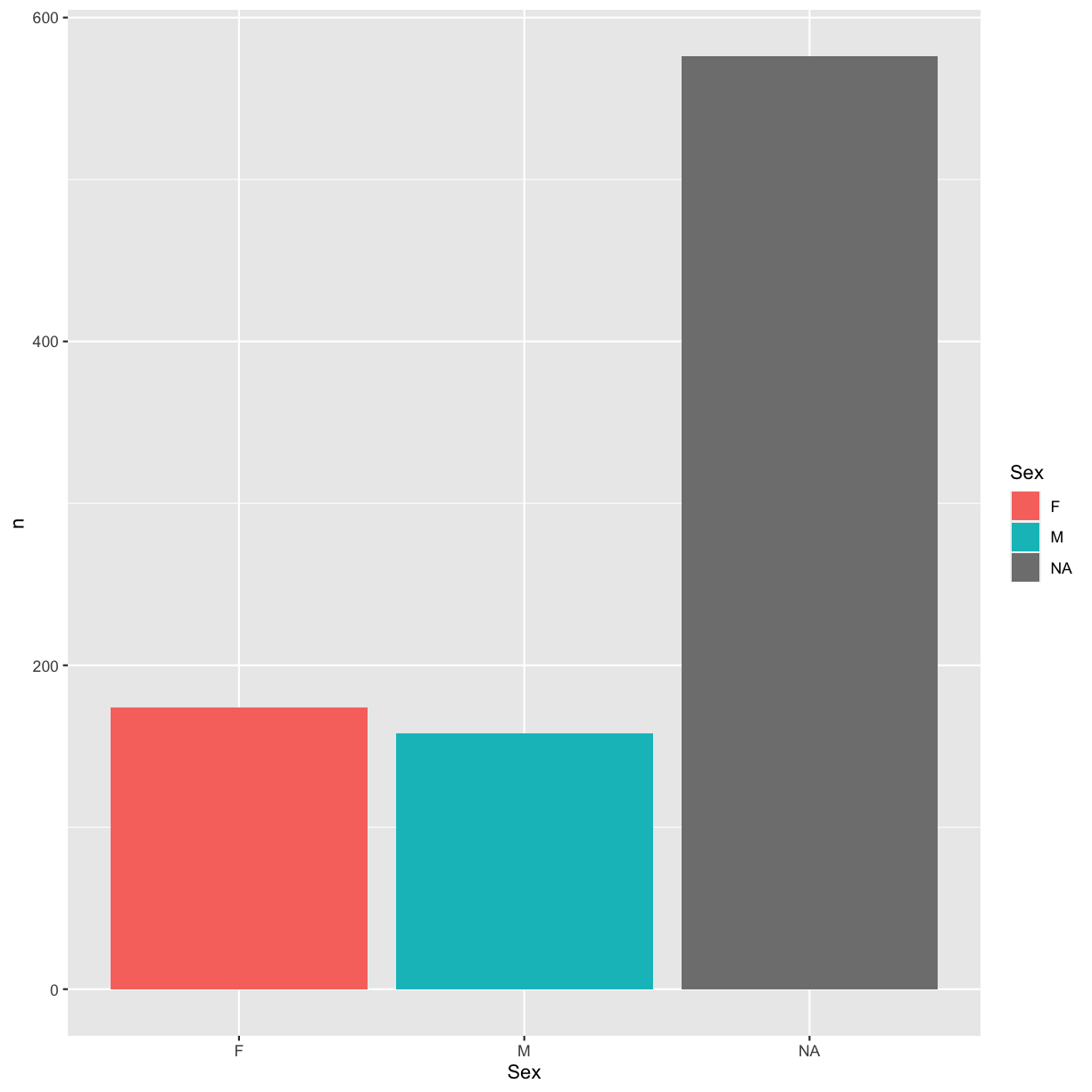
We could also have added this configuration to the geom_bar() layer instead:
ggplot(sex_counts, aes(x = Sex, y = n)) +
geom_bar(stat = "identity", aes(fill = Sex))
Plotting time series data
Let’s calculate number of counts per year for the Cooper’s and Sharp-shinned hawks. First we need to group the data and count records within each group.
yearly_sp_counts <- hawks %>%
filter(Species == "CH" | Species == "SS") %>%
count(Year, Species)
Timelapse data can be visualized as a line plot with years on the x-axis and counts on the y-axis:
ggplot(data = yearly_sp_counts, aes(x = Year, y = n)) +
geom_line()
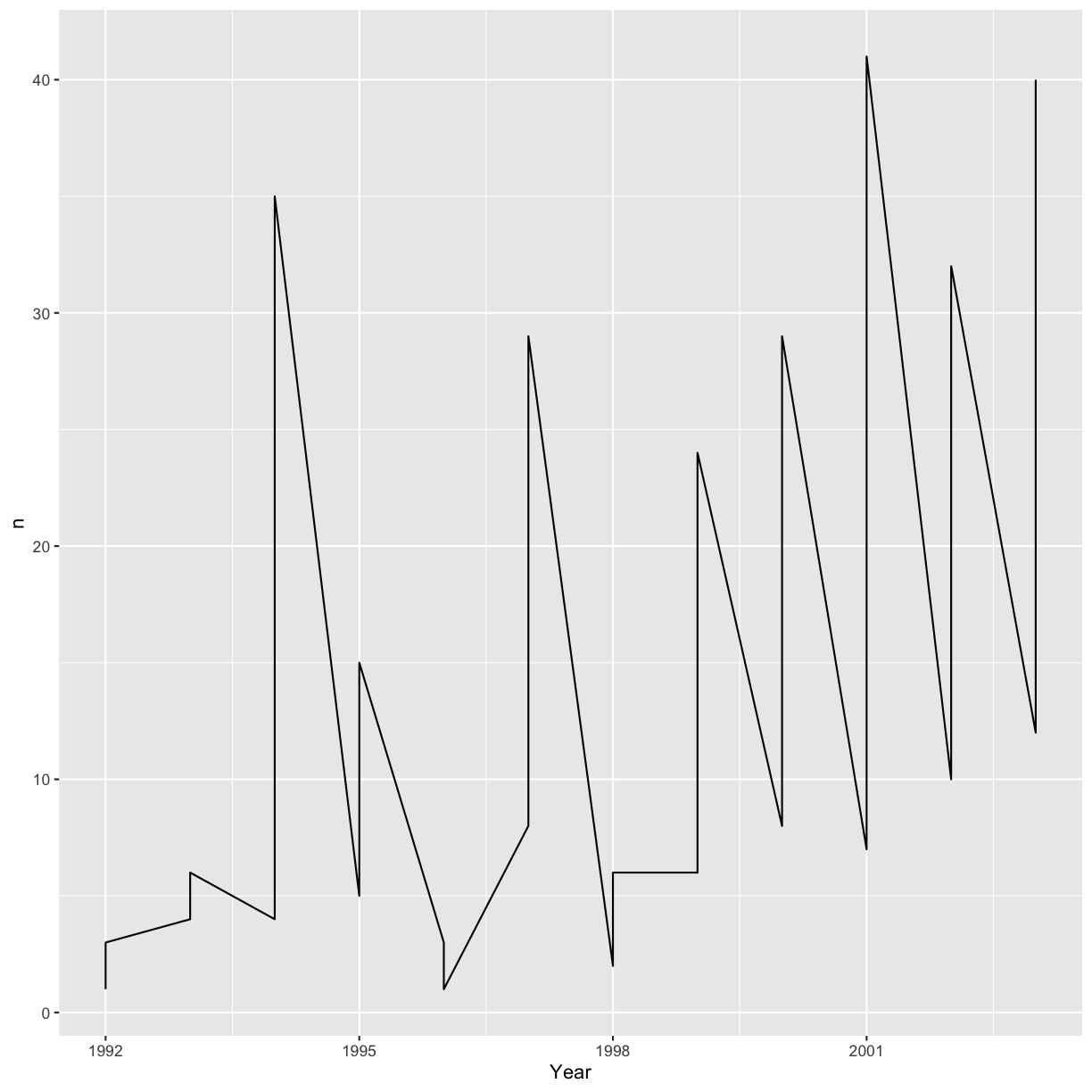
Unfortunately, this does not work because we plotted data for both the species together. We need to tell ggplot to draw a line for each species by modifying the aesthetic function to include group = Species:
ggplot(data = yearly_sp_counts, aes(x = Year, y = n, group = Species)) +
geom_line()
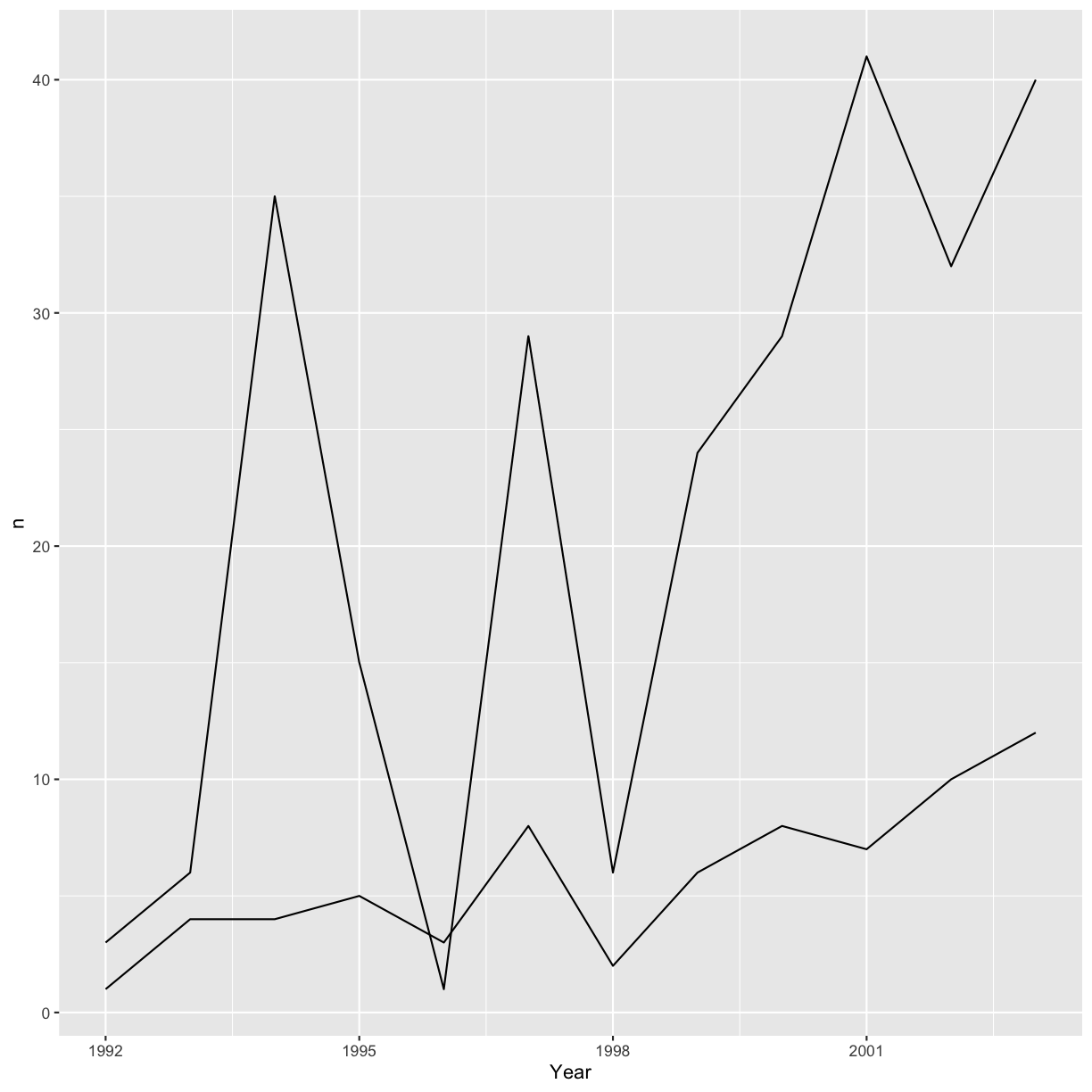
We will be able to distinguish the species in the plot if we add colors (using
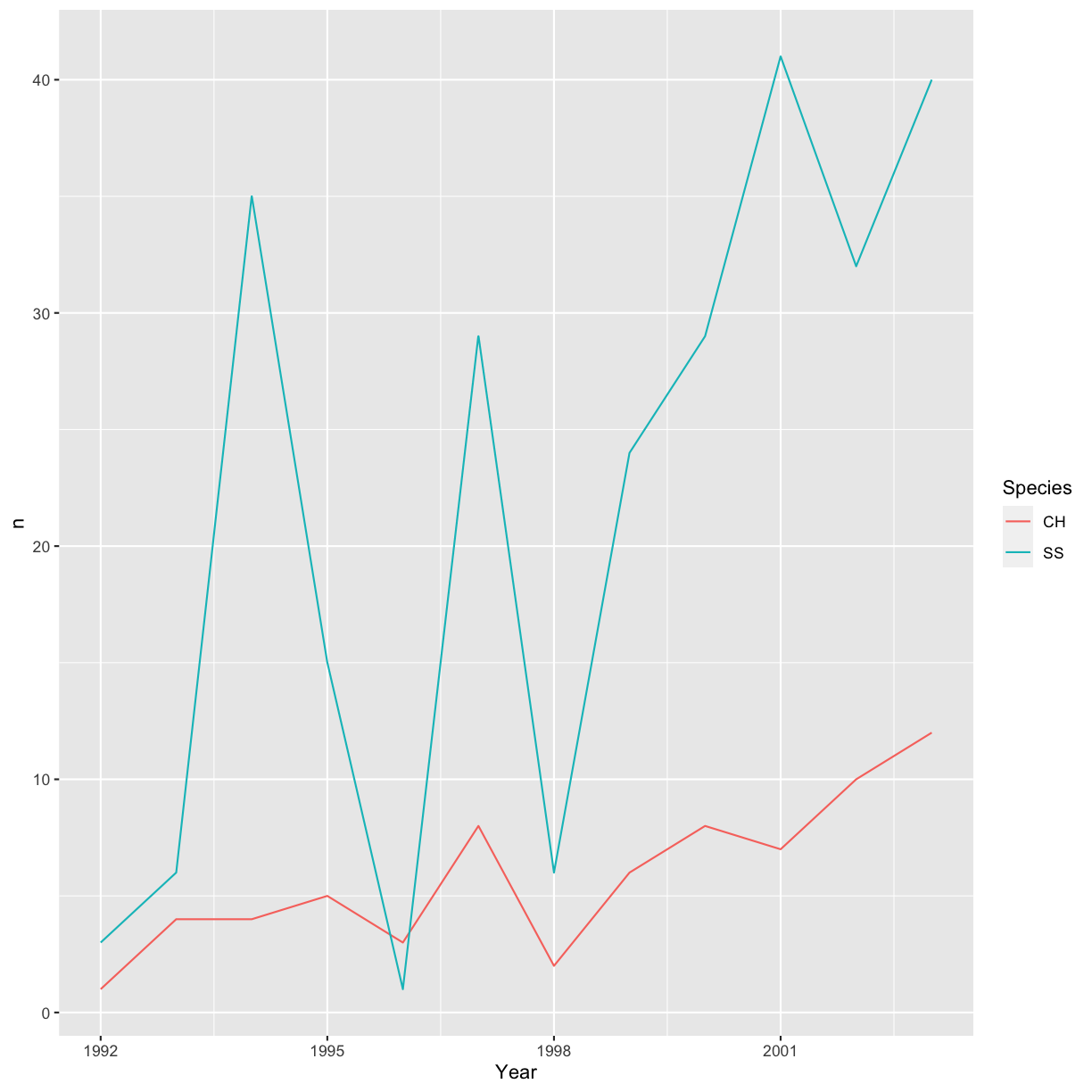
color also automatically groups the data):
ggplot(data = yearly_sp_counts, aes(x = Year, y = n, color = Species)) +
geom_line()
Integrating the pipe operator with ggplot2
In the previous lesson, we saw how to use the pipe operator %>% to use
different functions in a sequence and create a coherent workflow.
We can also use the pipe operator to pass the data argument to the
ggplot() function. The hard part is to remember that to build your ggplot,
you need to use + and not %>%.
yearly_sp_counts %>% ggplot(aes(x = Year, y = n, color = Species)) +
geom_line()
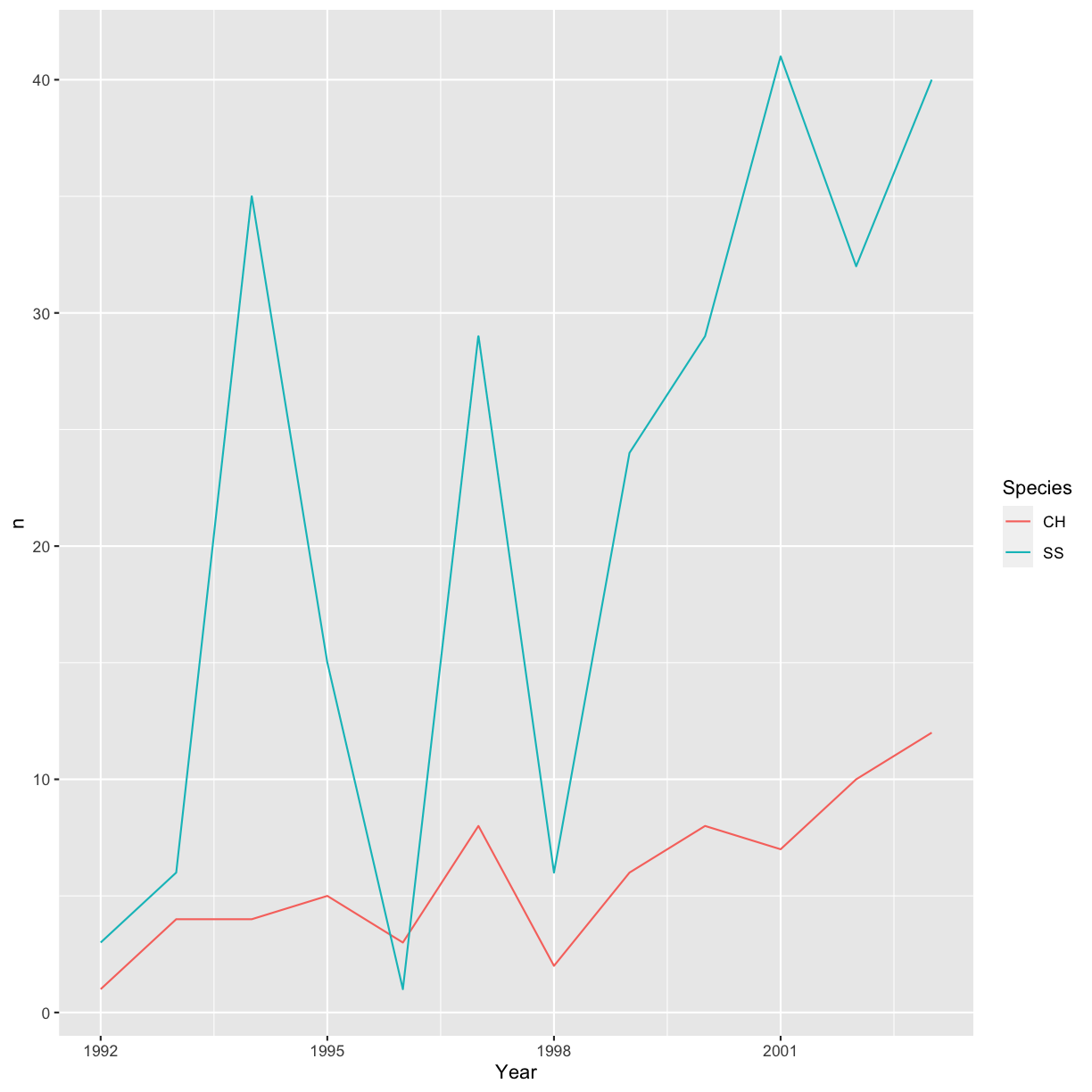
The pipe operator can also be used to link data manipulation with consequent data visualization.
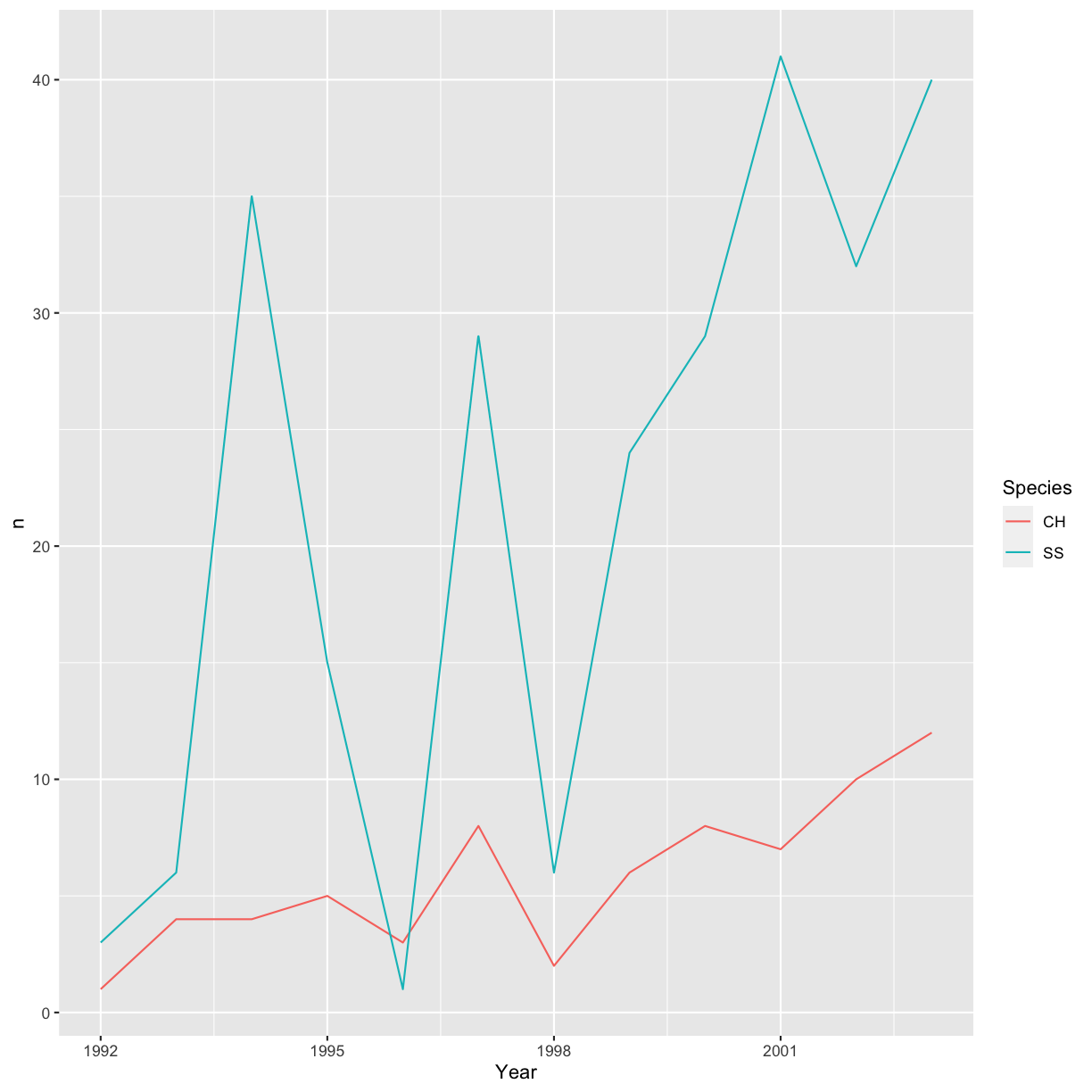
yearly_sp_plot <- hawks %>%
filter(Species == "CH" | Species == "SS") %>%
count(Year, Species) %>%
ggplot(aes(x = Year, y = n, color = Species)) +
geom_line()
yearly_sp_plot
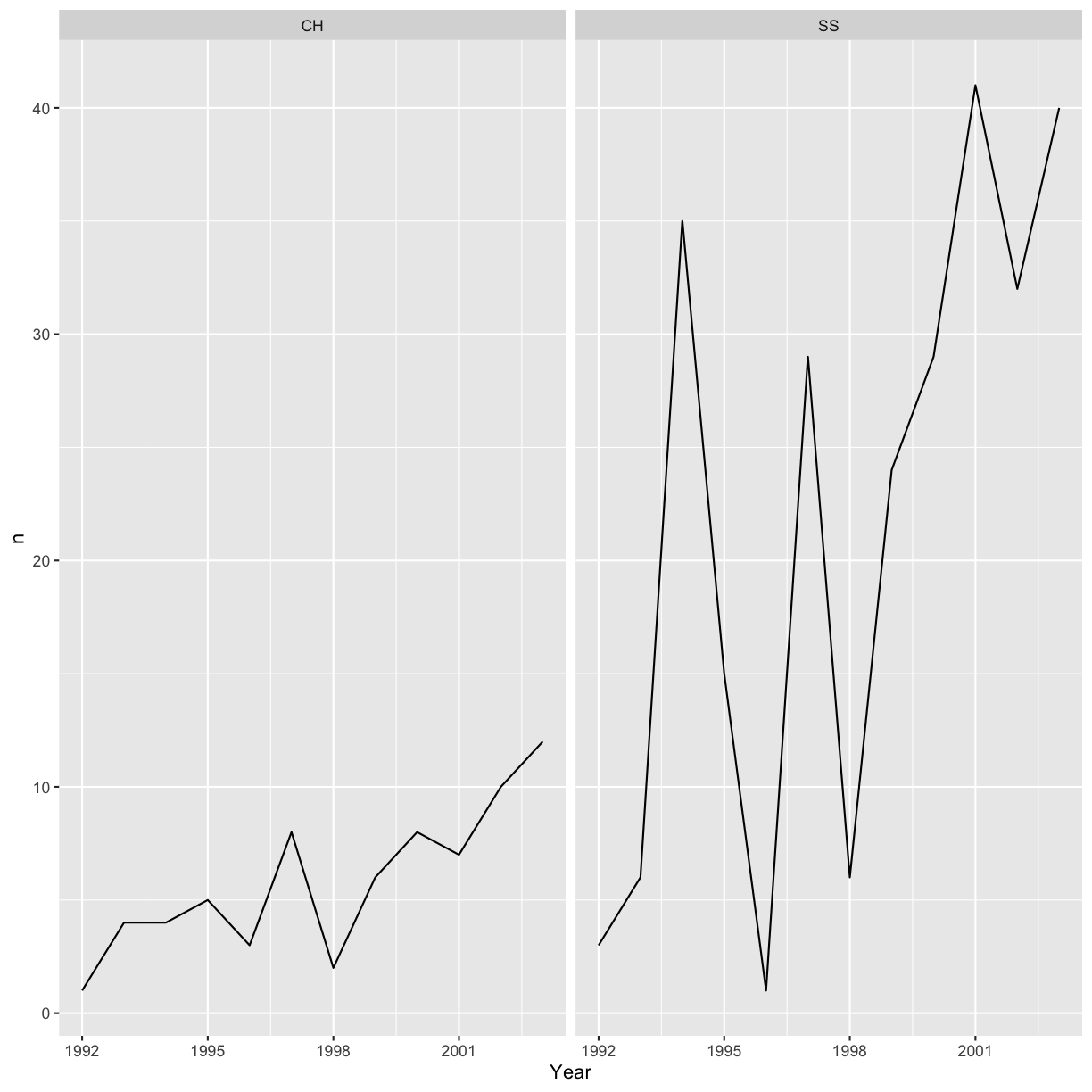
Faceting
ggplot has a special technique called faceting that allows the user to split
one plot into multiple plots based on a factor included in the dataset. We will
use it to make one lineplot for each of the species:
ggplot(data = yearly_sp_counts, aes(x = Year, y = n)) +
geom_line() +
facet_wrap(facets = vars(Species))
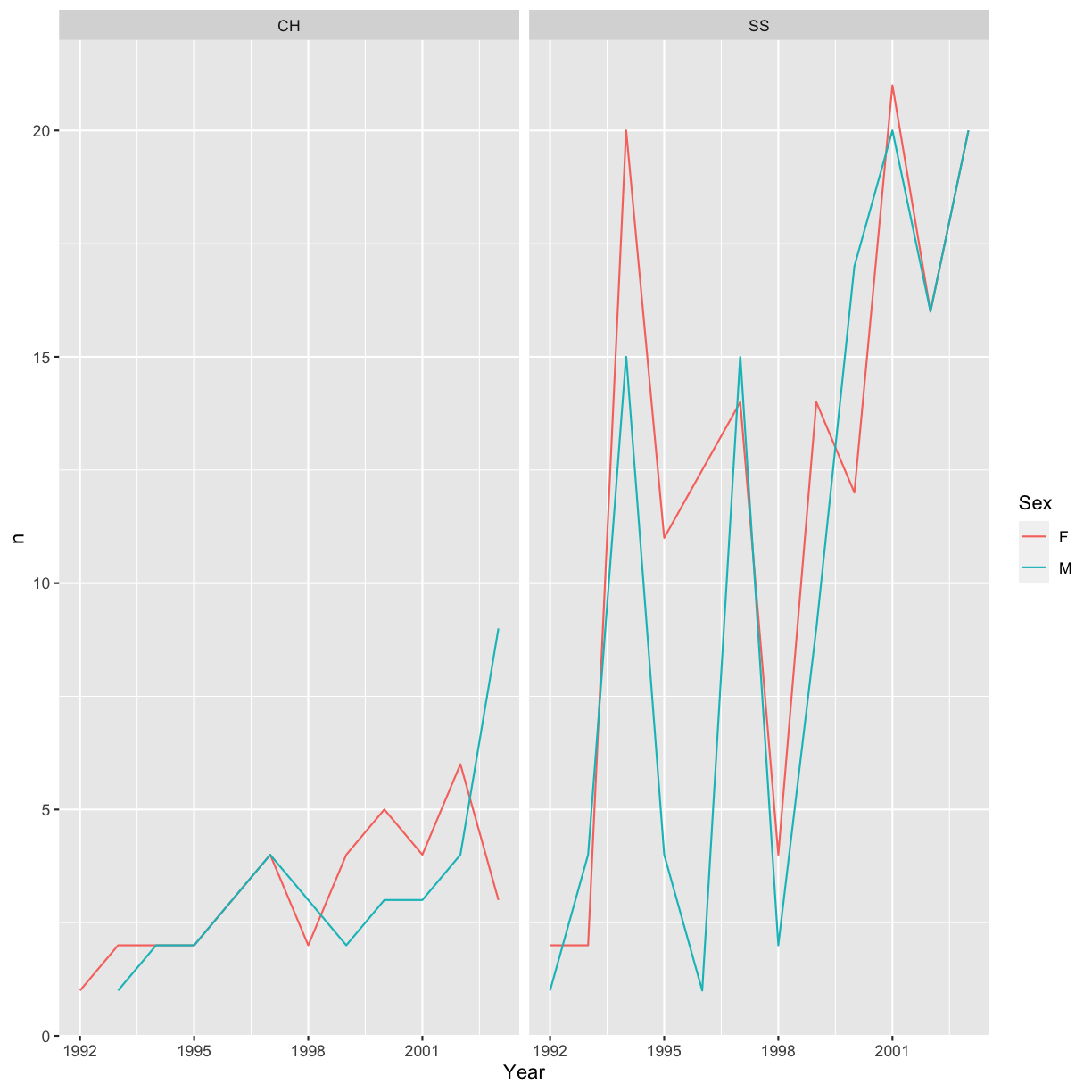
 Now we would like to split the line in each plot by the sex of each individual
measured. To do that we need to make counts in the data frame grouped by
Now we would like to split the line in each plot by the sex of each individual
measured. To do that we need to make counts in the data frame grouped by Year,
Species, and Sex. We will also drop rows with NA in any of these three columns.
yearly_sp_sex_counts <- hawks %>%
filter(Species == "CH" | Species == "SS") %>%
drop_na(Year, Species, Sex) %>%
count(Year, Species, Sex)
We can now make the faceted plot by splitting further by sex using color
(within a single plot):
ggplot(data = yearly_sp_sex_counts, aes(x = Year, y = n, color = Sex)) +
geom_line() +
facet_wrap(facets = vars(Species))
It is also possible to create more advanced layouts using the facet_grid()
function. We can for example facet both by species and sex:
ggplot(data = yearly_sp_sex_counts, aes(x = Year, y = n, color = Sex, group = Sex)) +
geom_line() +
facet_grid(rows = vars(Sex), cols = vars(Species))
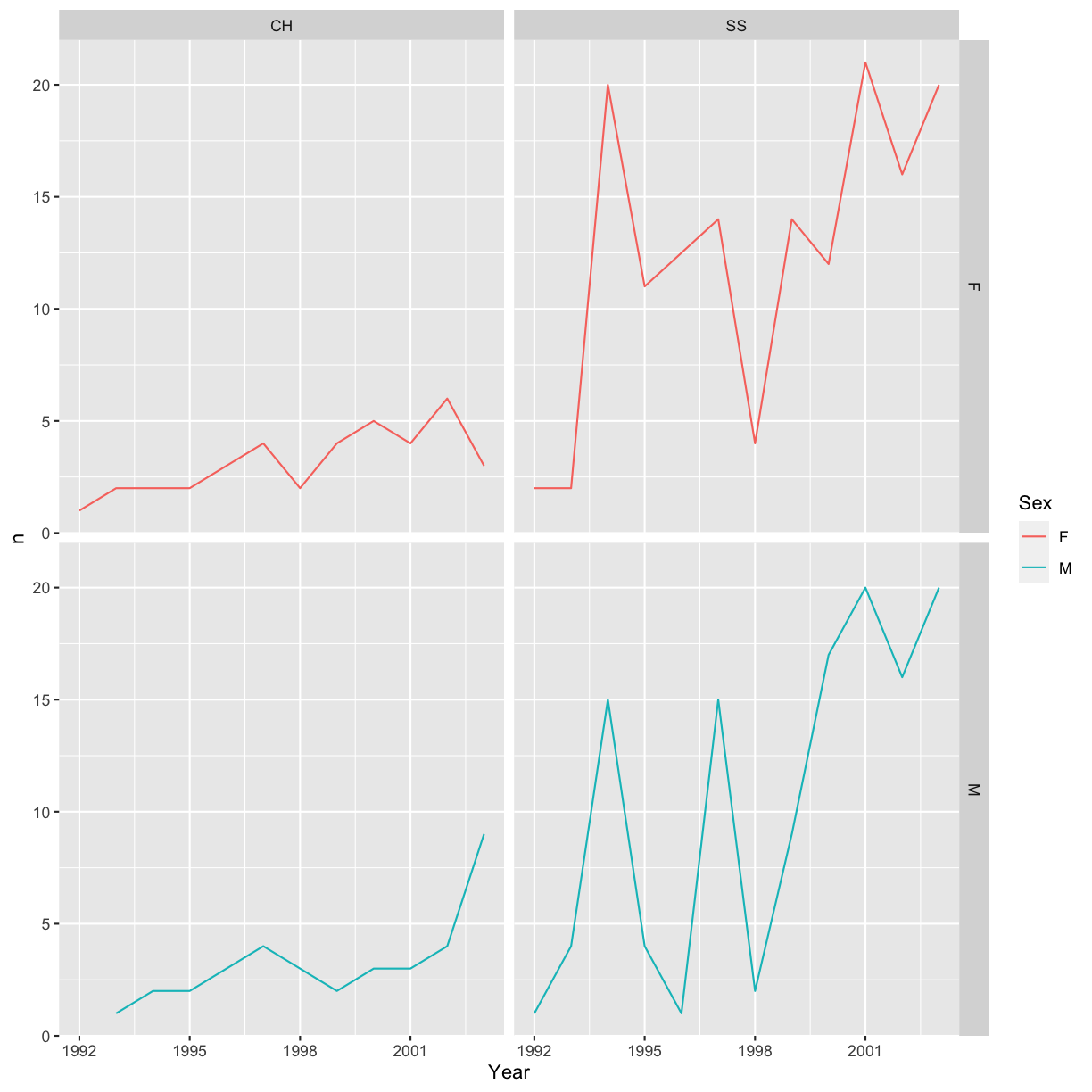
You can also organise the panels only by rows (or only by columns):
# One row, facet by column
ggplot(data = yearly_sp_sex_counts, aes(x = Year, y = n, color = Sex)) +
geom_line() +
facet_grid(rows = vars(Species))
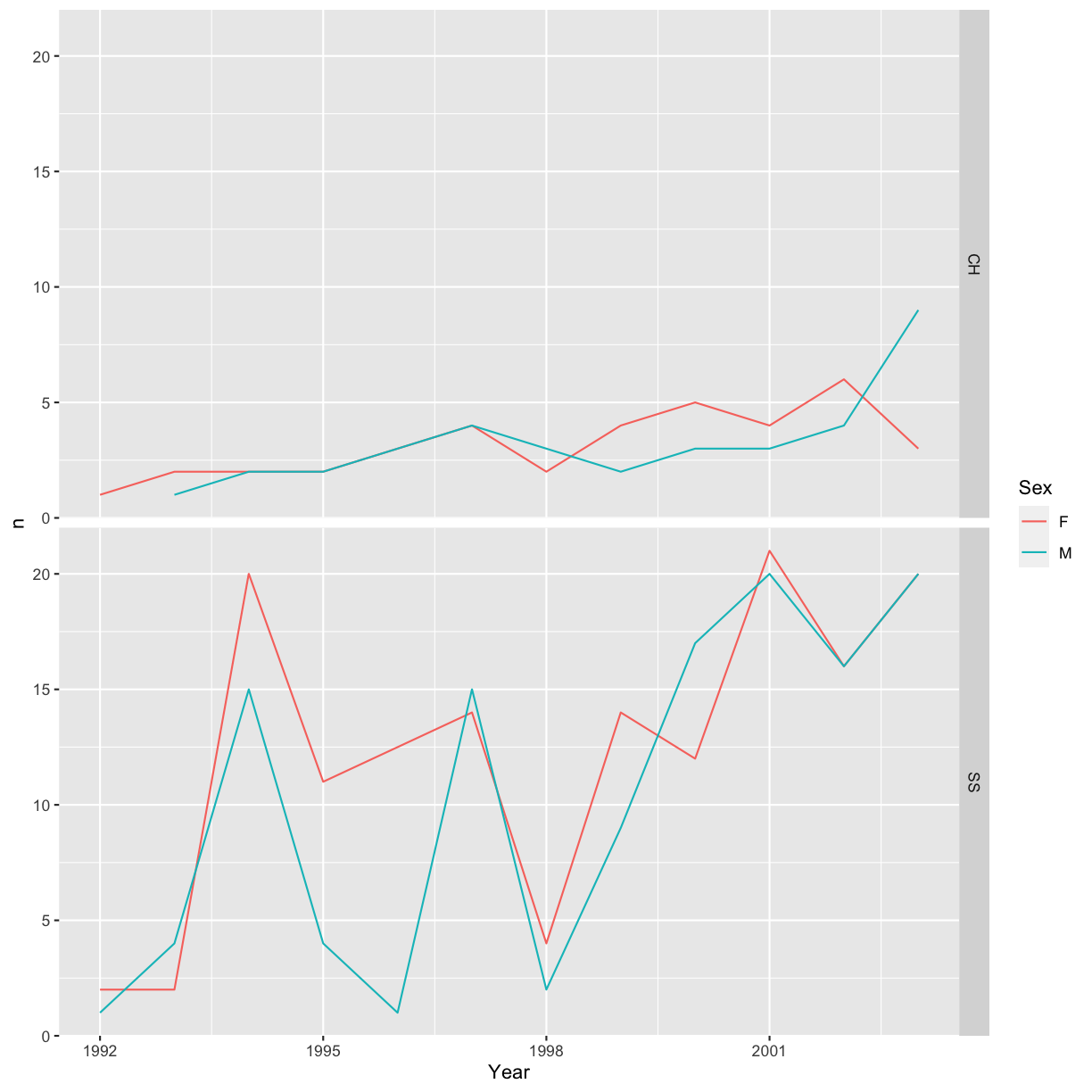
Note
ggplot2before version 3.0.0 used formulas to specify how plots are faceted. If you encounterfacet_grid/wrap(...)code containing~, please read https://ggplot2.tidyverse.org/news/#ggplot2-300.
ggplot2 themes
Usually plots with white background look more readable when printed. Every
single component of a ggplot graph can be customized using the generic
theme() function, as we will see below. However, there are pre-loaded themes
available that change the overall appearance of the graph without much effort.
For example, we can change our previous graph to have a simpler white background
using the theme_bw() function:
ggplot(data = yearly_sp_sex_counts, aes(x = Year, y = n, color = Sex)) +
geom_line() +
facet_wrap(facets = vars(Species)) +
theme_bw()
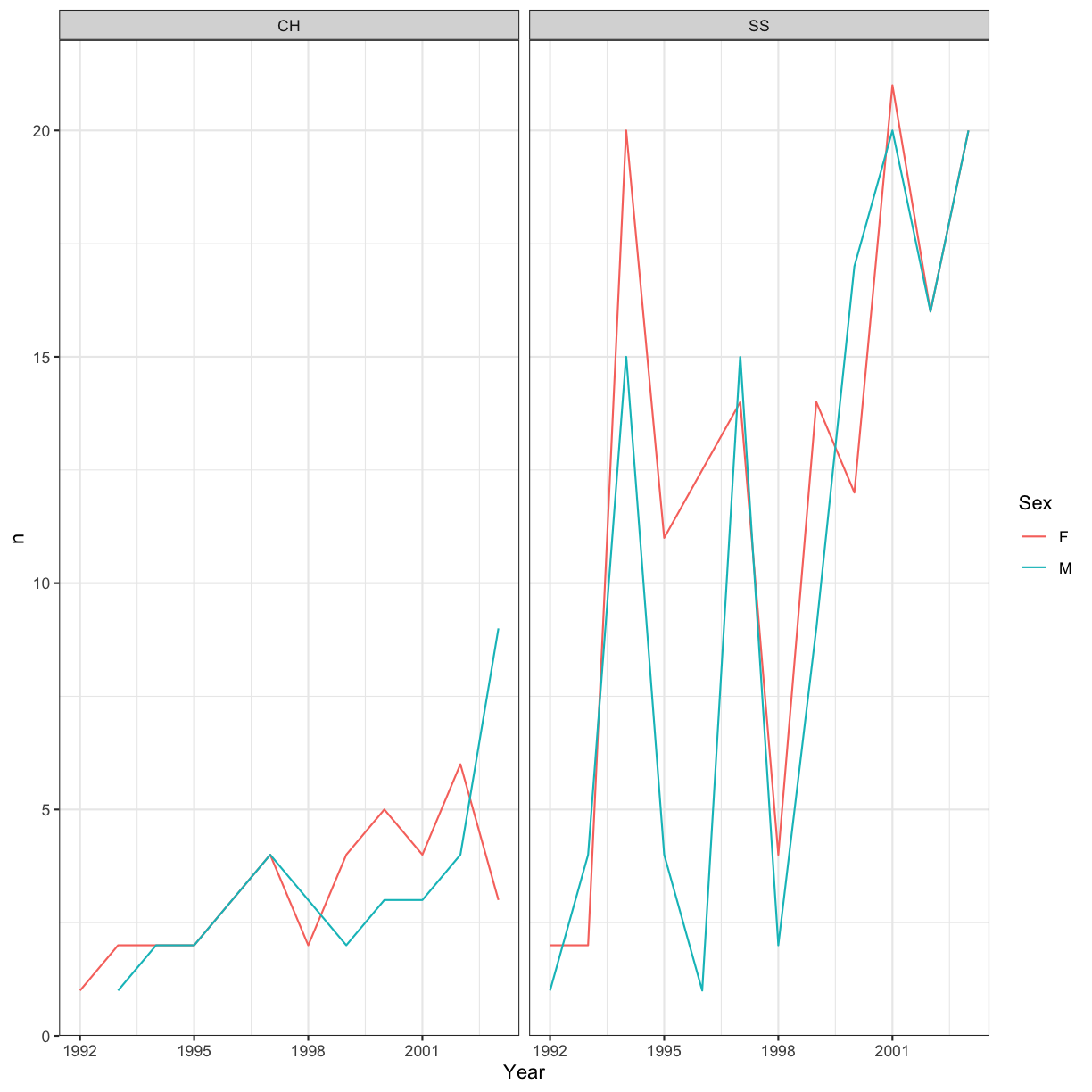
In addition to theme_bw(), which changes the plot background to white,
ggplot2 comes with several other themes which can be useful to quickly
change the look of your visualization. The complete list of themes is available
at https://ggplot2.tidyverse.org/reference/ggtheme.html. theme_minimal() and
theme_light() are popular, and theme_void() can be useful as a starting
point to create a new hand-crafted theme.
The ggthemes package provides a wide variety of options.
Challenge 4.4
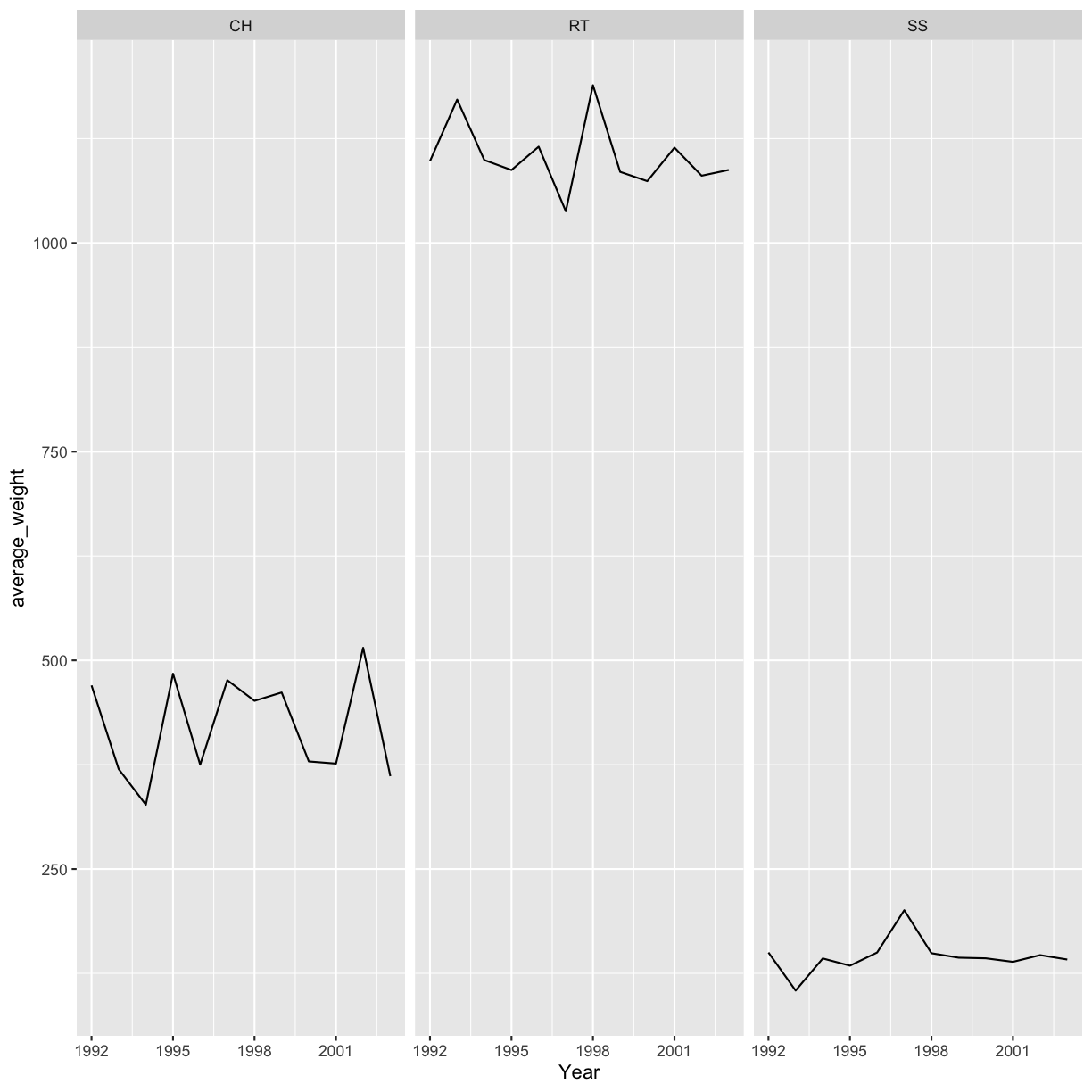
Use what you just learned to create a plot that depicts how the average weight of each species changes through the years.
Solution
yearly_weight <- hawks %>% group_by(Species, Year) %>% summarize(average_weight = mean(Weight, na.rm = TRUE)) ggplot(data = yearly_weight, aes(x=Year, y = average_weight)) + geom_line() + facet_wrap(vars(Species))
Customization
Take a look at the ggplot2 cheat sheet, and
think of ways you could improve the plot.
Now, let’s start with changing the names of axes and add a title to the figure:
ggplot(data = yearly_sp_sex_counts, aes(x = Year, y = n, color = Sex)) +
geom_line() +
facet_wrap(vars(Species)) +
labs(title = "Captured individuals through time",
x = "Year of capture",
y = "Number of individuals") +
theme_bw()
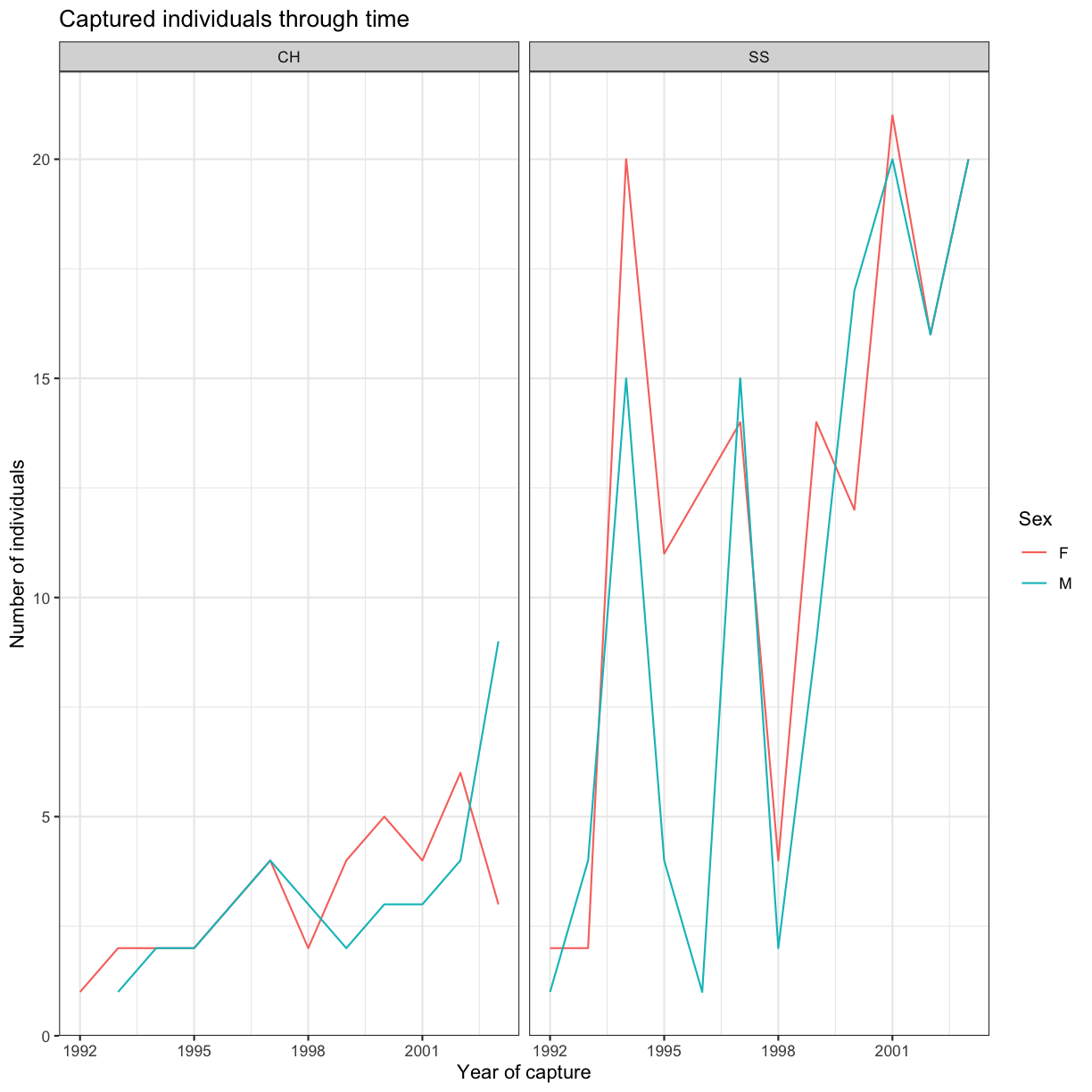
The axes have more informative names, but their readability can be improved by
increasing the font size. This can be done with the generic theme() function:
ggplot(data = yearly_sp_sex_counts, aes(x = Year, y = n, color = Sex)) +
geom_line() +
facet_wrap(vars(Species)) +
labs(title = "Captured individuals through time",
x = "Year of capture",
y = "Number of individuals") +
theme_bw() +
theme(text = element_text(size = 16)) # set the font size of text elements
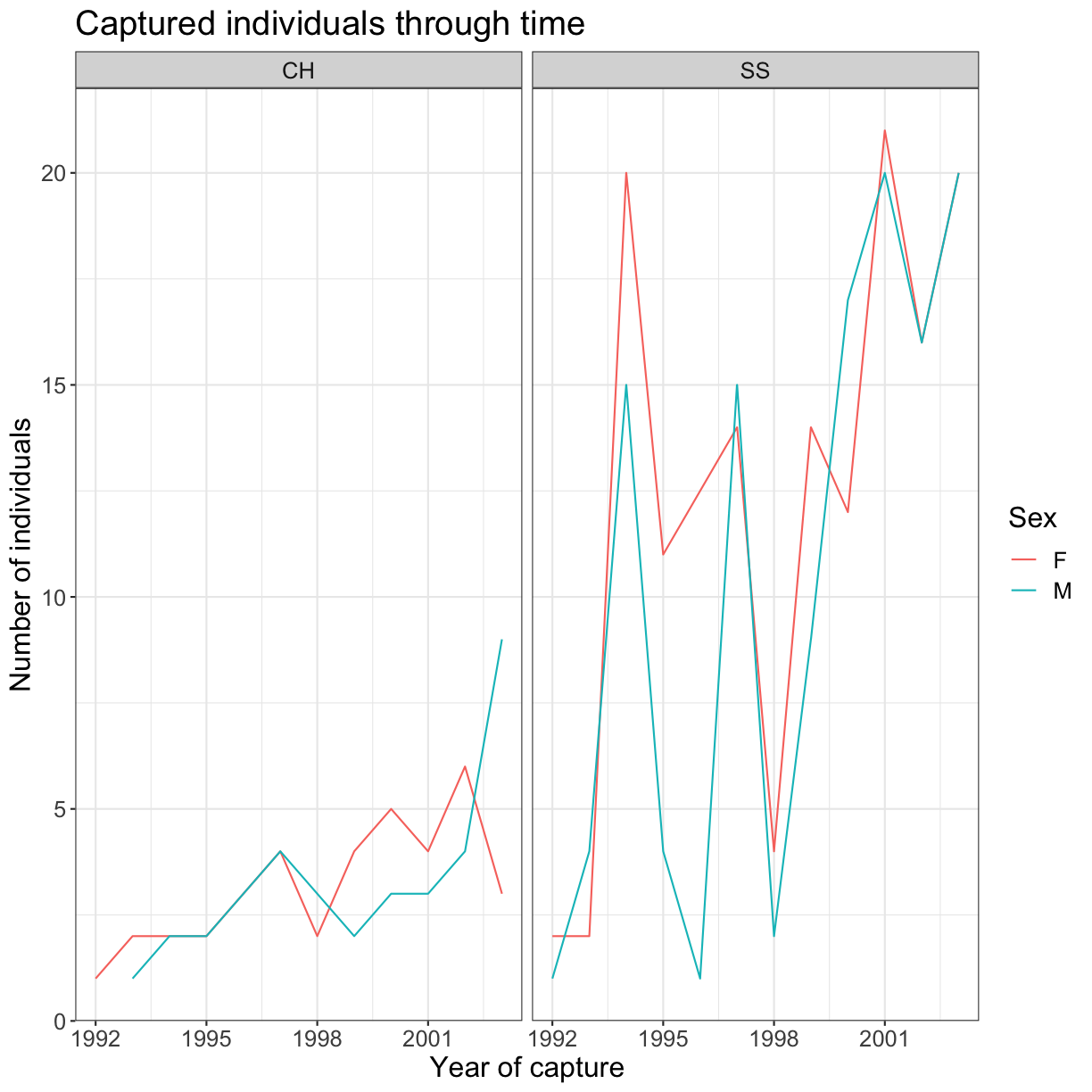
To alter the species and sex labels, we create factors and modify the levels:
yearly_sp_sex_counts <- yearly_sp_sex_counts %>% mutate(
Species = factor(Species),
Sex = factor(Sex)
)
levels(yearly_sp_sex_counts$Species)[1:2] = c("Cooper's hawk", "Sharp-shinned hawk")
levels(yearly_sp_sex_counts$Sex)[1:2] = c("Female", "Male")
Now, run the code to generate the plots once again:
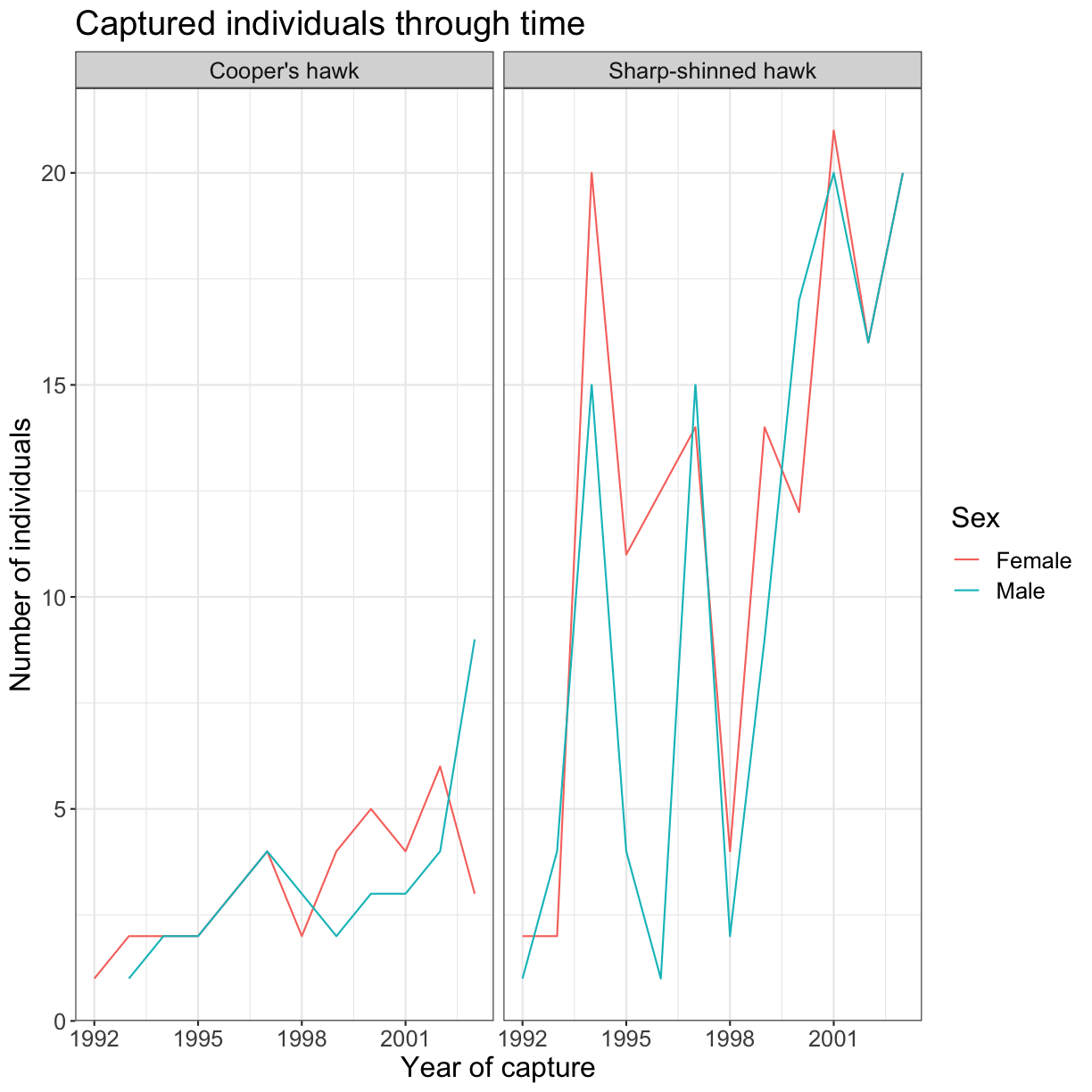
ggplot(data = yearly_sp_sex_counts, aes(x = Year, y = n, color = Sex)) +
geom_line() +
facet_wrap(vars(Species)) +
labs(
title = "Captured individuals through time",
x = "Year of capture",
y = "Number of individuals") +
theme_bw() +
theme(text = element_text(size = 16))
Note that it is also possible to change the fonts of your plots. If you are on
Windows, you may have to install the extrafont package,
and follow the instructions included in the README for this package.
Challenge 4.5
With all of this information in hand, please take another five minutes to either improve one of the plots generated in this exercise or create a beautiful graph of your own. Use the RStudio
ggplot2cheat sheet for inspiration.Here are some ideas:
- See if you can change the thickness of the lines.
- Can you find a way to change the name of the legend?
- Try using a different color palette (see http://www.cookbook-r.com/Graphs/Colors_(ggplot2)/).
Arranging plots
Faceting is a great tool for splitting one plot into multiple plots, but
sometimes you may want to produce a single figure that contains multiple plots
using different variables or even different data frames. We won’t go into
it here, but the * patchwork package can be used to combine separate
gplots into a single figure while keeping everything aligned properly. Like most
R packages, patchwork can be installed from CRAN, the R package repository.
Exporting plots
After creating your plot, you can save it to a file in your favorite format. The
Export tab in the Plot pane in RStudio will save your plots at low
resolution, which will not be accepted by many journals and will not scale well
for posters. The ggplot2 extensions website
provides a list of packages that extend the capabilities of ggplot2,
including additional themes.
Instead, use the ggsave() function, which allows you easily change the
dimension and resolution of your plot by adjusting the appropriate arguments
(width, height and dpi):
year_plot <- ggplot(data = yearly_sp_sex_counts, aes(x = Year, y = n, color = Sex)) +
geom_line() +
facet_wrap(vars(Species)) +
labs(
title = "Captured individuals through time",
x = "Year of capture",
y = "Number of individuals") +
theme_bw() +
theme(text = element_text(size = 16))
# Save the file to a subfolder named "figs"
ggsave("figs/year_plot.png", year_plot, width = 15, height = 10)