| avg_log2FC | p_val_adj | |
|---|---|---|
| CD7 | 5.535220 | 0.0000001 |
| LCK | 3.605886 | 0.0000046 |
| HLA-DPB1 | -5.291575 | 0.0000051 |
| HLA-DRA | -4.128576 | 0.0000126 |
| HLA-DRB1 | -5.027130 | 0.0000172 |
| GNLY | 8.198735 | 0.0000191 |
| GZMM | 3.120563 | 0.0000767 |
| CD3D | 2.255304 | 0.0000805 |
| GZMA | 3.078594 | 0.0001174 |
| HLA-DPA1 | -3.661491 | 0.0002595 |
Differential Gene Expression
Single Cell RNA-Seq Analysis
Jennifer Fransson
02-Apr-2025
Overview
- What is differential gene expression?
- How is the analysis performed?
- Choosing your groups of interest
- Functions
- Simple design vs complex design
- 1-vs-1 and 1-vs-all
- Other considerations
What is differential gene expression?
What is differential gene expression?

Count data -> statistical analysis -> Are differences significant (greater than expected randomly)
Statistical tests
- Null hypothesis: Mean/median/distribution is equal between group A and group B
- \(p\) < 0.05 : if null hypothesis is true, we can expect the measured result in < 5% of cases where group A and group B have been sampled with sample size \(n\)
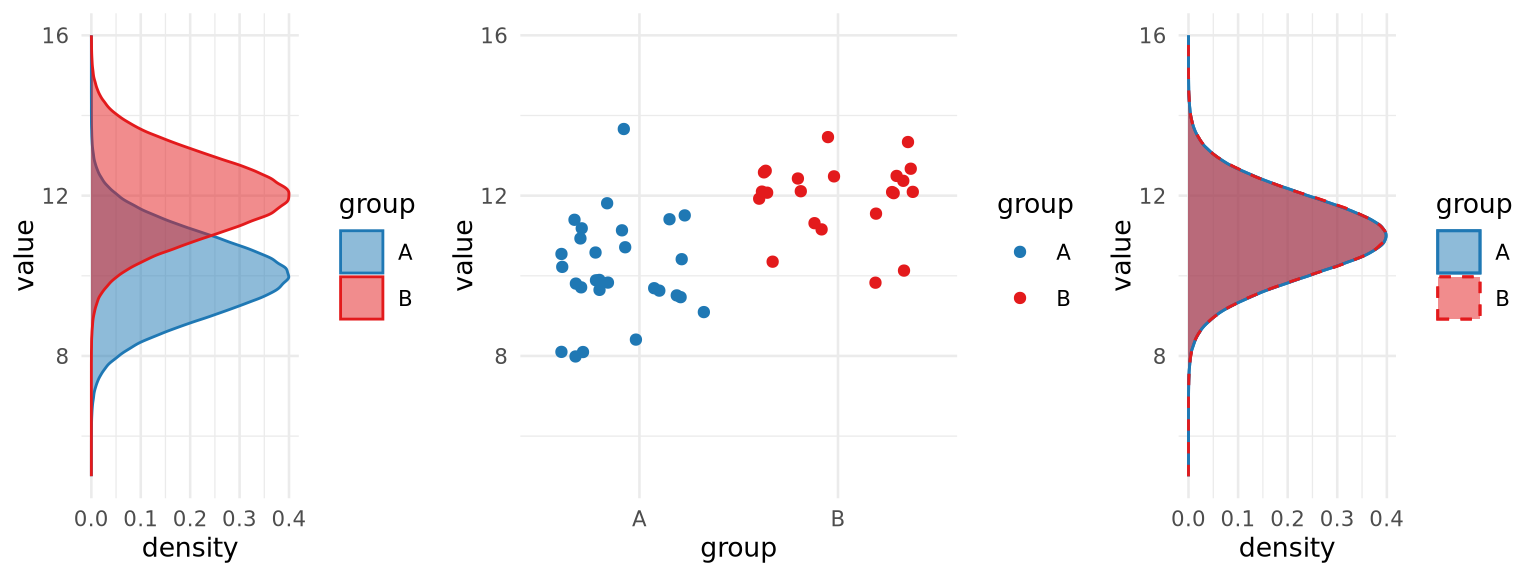
- Statistical significance: The result is likely to be from a true difference rather than random chance
What is differential gene expression?
What is differential gene expression?

What is differential gene expression?
| avg_log2FC | p_val_adj | pct.1 | pct.2 | |
|---|---|---|---|---|
| CD7 | 5.535220 | 0.0000001 | 0.714 | 0.058 |
| LCK | 3.605886 | 0.0000046 | 0.679 | 0.077 |
| HLA-DPB1 | -5.291575 | 0.0000051 | 0.107 | 0.769 |
| HLA-DRA | -4.128576 | 0.0000126 | 0.357 | 0.865 |
| HLA-DRB1 | -5.027130 | 0.0000172 | 0.107 | 0.731 |
| GNLY | 8.198735 | 0.0000191 | 0.571 | 0.058 |
| GZMM | 3.120563 | 0.0000767 | 0.571 | 0.058 |
| CD3D | 2.255304 | 0.0000805 | 0.643 | 0.096 |
| GZMA | 3.078594 | 0.0001174 | 0.571 | 0.058 |
| HLA-DPA1 | -3.661491 | 0.0002595 | 0.179 | 0.712 |
What is differential gene expression?

(Modified from Tiberi et al., 2023)
- Most methods focus on difference in mean
- Many different distributions will show a difference in means
- But not all!
How is the analysis performed?
Defining groups of interest
A priori defined groups: Compare cells from different samples, e.g.:
- Experimental groups (treatment, time points, clinical information etc)
- Sorted cells
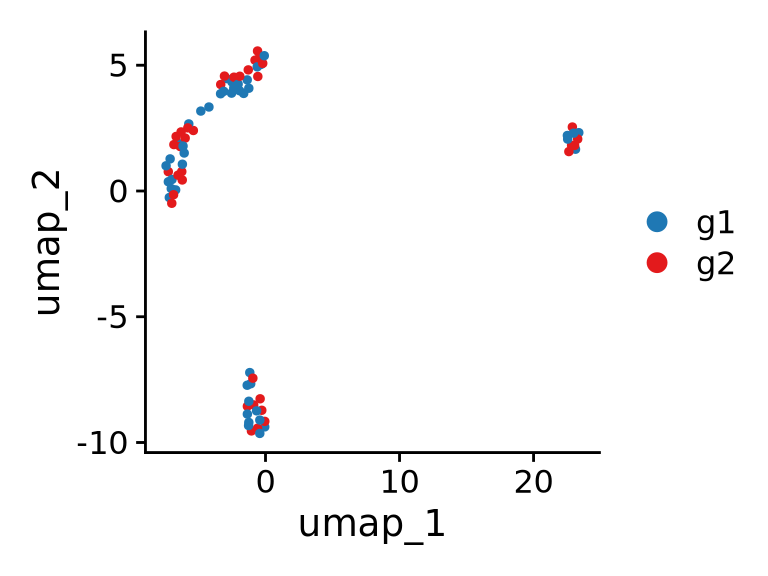
Data-driven definition of groups: Compare cells depending on analysis output, e.g.:
- RNA-based clustering/identity
- Identity based on other data from multi-omics
Warning: Performing DE on clusters defined with the same data (“double-dipping”) will inflate DE analysis. Be mindful of this when you interpret the results.
Functions
| Toolkit | Function |
|---|---|
| Seurat | FindMarkers(), FindAllMarkers() |
| Scran | findMarkers() |
| Scanpy | scanpy.tl.rank_genes_groups() |
FindAllMarkers
FindAllMarkers(
object,
assay = NULL,
features = NULL,
logfc.threshold = 0.1,
test.use = "wilcox",
slot = "data",
min.pct = 0.01,
min.diff.pct = -Inf,
node = NULL,
verbose = TRUE,
only.pos = FALSE,
max.cells.per.ident = Inf,
random.seed = 1,
latent.vars = NULL,
min.cells.feature = 3,
min.cells.group = 3,
mean.fxn = NULL,
fc.name = NULL,
base = 2,
return.thresh = 0.01,
densify = FALSE,
...
)Seurat 5.1.0
FindAllMarkers
FindAllMarkers(
object,
assay = NULL,
features = NULL,
logfc.threshold = 0.1,
test.use = "wilcox",
slot = "data",
min.pct = 0.01,
min.diff.pct = -Inf,
node = NULL,
verbose = TRUE,
only.pos = FALSE,
max.cells.per.ident = Inf,
random.seed = 1,
latent.vars = NULL,
min.cells.feature = 3,
min.cells.group = 3,
mean.fxn = NULL,
fc.name = NULL,
base = 2,
return.thresh = 0.01,
densify = FALSE,
...
)Seurat 5.1.0
FindAllMarkers
FindAllMarkers(
object,
assay = NULL,
features = NULL,
logfc.threshold = 0.1,
test.use = "wilcox",
slot = "data",
min.pct = 0.01,
min.diff.pct = -Inf,
node = NULL,
verbose = TRUE,
only.pos = FALSE,
max.cells.per.ident = Inf,
random.seed = 1,
latent.vars = NULL,
min.cells.feature = 3,
min.cells.group = 3,
mean.fxn = NULL,
fc.name = NULL,
base = 2,
return.thresh = 0.01,
densify = FALSE,
...
)Seurat 5.1.0
FindAllMarkers
FindAllMarkers(
object,
assay = NULL,
features = NULL,
logfc.threshold = 0.1,
test.use = "wilcox",
slot = "data",
min.pct = 0.01,
min.diff.pct = -Inf,
node = NULL,
verbose = TRUE,
only.pos = FALSE,
max.cells.per.ident = Inf,
random.seed = 1,
latent.vars = NULL,
min.cells.feature = 3,
min.cells.group = 3,
mean.fxn = NULL,
fc.name = NULL,
base = 2,
return.thresh = 0.01,
densify = FALSE,
...
)Seurat 5.1.0
Statistical tests
"wilcox" : Identifies differentially expressed genes between two groups of cells using a Wilcoxon Rank Sum test (default)
"bimod" : Likelihood-ratio test for single cell gene expression, (McDavid et al., Bioinformatics, 2013)
"roc" : Identifies 'markers' of gene expression using ROC analysis. For each gene, evaluates (using AUC) a classifier built on that gene alone, to classify between two groups of cells. An AUC value of 1 means that expression values for this gene alone can perfectly classify the two groupings (i.e. Each of the cells in cells.1 exhibit a higher level than each of the cells in cells.2). An AUC value of 0 also means there is perfect classification, but in the other direction. A value of 0.5 implies that the gene has no predictive power to classify the two groups. Returns a 'predictive power' (abs(AUC-0.5) * 2) ranked matrix of putative differentially expressed genes.
"t" : Identify differentially expressed genes between two groups of cells using the Student's t-test.
"negbinom" : Identifies differentially expressed genes between two groups of cells using a negative binomial generalized linear model. Use only for UMI-based datasets
"poisson" : Identifies differentially expressed genes between two groups of cells using a poisson generalized linear model. Use only for UMI-based datasets
"LR" : Uses a logistic regression framework to determine differentially expressed genes. Constructs a logistic regression model predicting group membership based on each feature individually and compares this to a null model with a likelihood ratio test.
"MAST" : Identifies differentially expressed genes between two groups of cells using a hurdle model tailored to scRNA-seq data. Utilizes the MAST package to run the DE testing.
"DESeq2" : Identifies differentially expressed genes between two groups of cells based on a model using DESeq2 which uses a negative binomial distribution (Love et al, Genome Biology, 2014).This test does not support pre-filtering of genes based on average difference (or percent detection rate) between cell groups. However, genes may be pre-filtered based on their minimum detection rate (min.pct) across both cell groups. To use this method, please install DESeq2, using the instructions at https://bioconductor.org/packages/release/bioc/html/DESeq2.htmlStatistical tests
"wilcox" : Identifies differentially expressed genes between two groups of cells using a Wilcoxon Rank Sum test (default)
"bimod" : Likelihood-ratio test for single cell gene expression, (McDavid et al., Bioinformatics, 2013)
"roc" : Identifies 'markers' of gene expression using ROC analysis. For each gene, evaluates (using AUC) a classifier built on that gene alone, to classify between two groups of cells. An AUC value of 1 means that expression values for this gene alone can perfectly classify the two groupings (i.e. Each of the cells in cells.1 exhibit a higher level than each of the cells in cells.2). An AUC value of 0 also means there is perfect classification, but in the other direction. A value of 0.5 implies that the gene has no predictive power to classify the two groups. Returns a 'predictive power' (abs(AUC-0.5) * 2) ranked matrix of putative differentially expressed genes.
"t" : Identify differentially expressed genes between two groups of cells using the Student's t-test.
"negbinom" : Identifies differentially expressed genes between two groups of cells using a negative binomial generalized linear model. Use only for UMI-based datasets
"poisson" : Identifies differentially expressed genes between two groups of cells using a poisson generalized linear model. Use only for UMI-based datasets
"LR" : Uses a logistic regression framework to determine differentially expressed genes. Constructs a logistic regression model predicting group membership based on each feature individually and compares this to a null model with a likelihood ratio test.
"MAST" : Identifies differentially expressed genes between two groups of cells using a hurdle model tailored to scRNA-seq data. Utilizes the MAST package to run the DE testing.
"DESeq2" : Identifies differentially expressed genes between two groups of cells based on a model using DESeq2 which uses a negative binomial distribution (Love et al, Genome Biology, 2014).This test does not support pre-filtering of genes based on average difference (or percent detection rate) between cell groups. However, genes may be pre-filtered based on their minimum detection rate (min.pct) across both cell groups. To use this method, please install DESeq2, using the instructions at https://bioconductor.org/packages/release/bioc/html/DESeq2.htmlDistributions
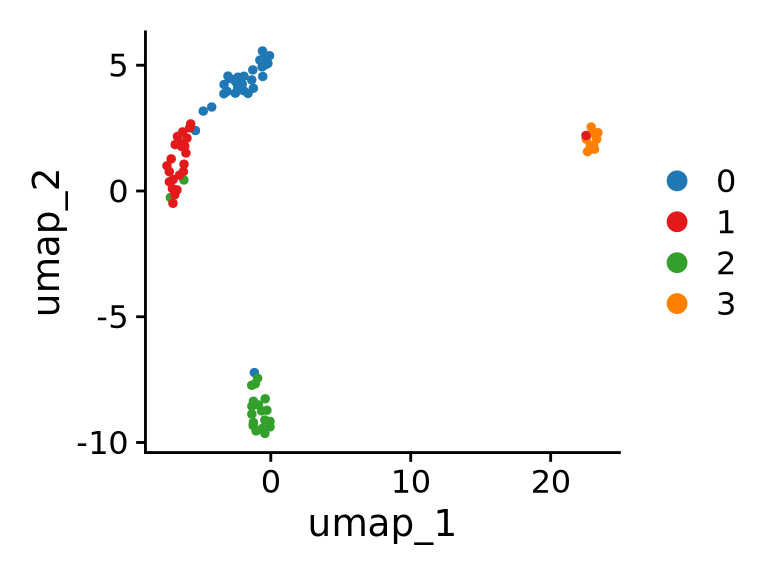
- High noise (technical + biology)
- Low library sizes
- Low mRNA quantity
- Amplification bias, drop-outs
- 3’ bias, partial coverage
- Bursting
- Mixed cell types
Statistical tests
"wilcox" : Identifies differentially expressed genes between two groups of cells using a Wilcoxon Rank Sum test (default)
"bimod" : Likelihood-ratio test for single cell gene expression, (McDavid et al., Bioinformatics, 2013)
"roc" : Identifies 'markers' of gene expression using ROC analysis. For each gene, evaluates (using AUC) a classifier built on that gene alone, to classify between two groups of cells. An AUC value of 1 means that expression values for this gene alone can perfectly classify the two groupings (i.e. Each of the cells in cells.1 exhibit a higher level than each of the cells in cells.2). An AUC value of 0 also means there is perfect classification, but in the other direction. A value of 0.5 implies that the gene has no predictive power to classify the two groups. Returns a 'predictive power' (abs(AUC-0.5) * 2) ranked matrix of putative differentially expressed genes.
"t" : Identify differentially expressed genes between two groups of cells using the Student's t-test.
"negbinom" : Identifies differentially expressed genes between two groups of cells using a negative binomial generalized linear model. Use only for UMI-based datasets
"poisson" : Identifies differentially expressed genes between two groups of cells using a poisson generalized linear model. Use only for UMI-based datasets
"LR" : Uses a logistic regression framework to determine differentially expressed genes. Constructs a logistic regression model predicting group membership based on each feature individually and compares this to a null model with a likelihood ratio test.
"MAST" : Identifies differentially expressed genes between two groups of cells using a hurdle model tailored to scRNA-seq data. Utilizes the MAST package to run the DE testing.
"DESeq2" : Identifies differentially expressed genes between two groups of cells based on a model using DESeq2 which uses a negative binomial distribution (Love et al, Genome Biology, 2014).This test does not support pre-filtering of genes based on average difference (or percent detection rate) between cell groups. However, genes may be pre-filtered based on their minimum detection rate (min.pct) across both cell groups. To use this method, please install DESeq2, using the instructions at https://bioconductor.org/packages/release/bioc/html/DESeq2.htmlHurdle models
…most computational methods still stick with the old mentality of viewing differential expression as a simple ‘up or down’ phenomenon. We advocate that we should fully embrace the features of single cell data, which allows us to observe binary (from Off to On) as well as continuous (the amount of expression) regulations. Wu et al. (2018)
MAST
- Two part GLM (Hurdle model)
- Models the continuous nature of gene expression and the discrete binary nature of gene detection
- Detection hurdle
- Expression detected or not?
- Logistic regression
- If gene is not detected, stop, else move to next hurdle
- Expression hurdle
- Genes with positive expression levels modelled using GLM
- Hurdle model is able to handle drop-outs
- Support complex modelling
Finak et al. (2015)
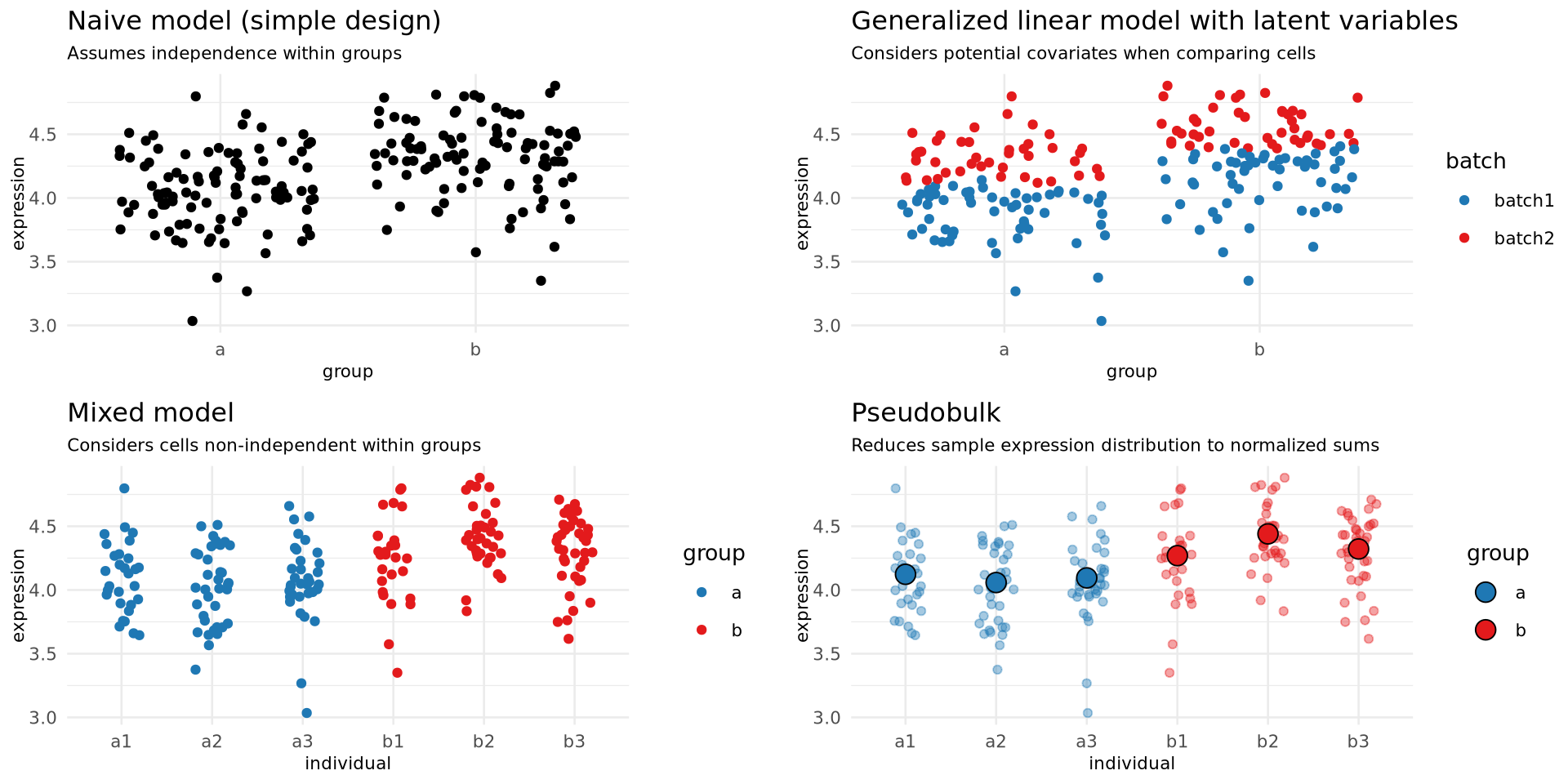
Complex designs
- Comparing groups of samples (e.g. patients vs controls)
- Including batch effects
- Correcting for covariates (e.g. age)
Complex designs: Groups of samples
Example: 3 patients vs 3 controls
\(n\): Number of cells (1000s) or number of individuals (3)?

Many tests assume independence!
Complex designs: Covariates
Complex designs: Covariates
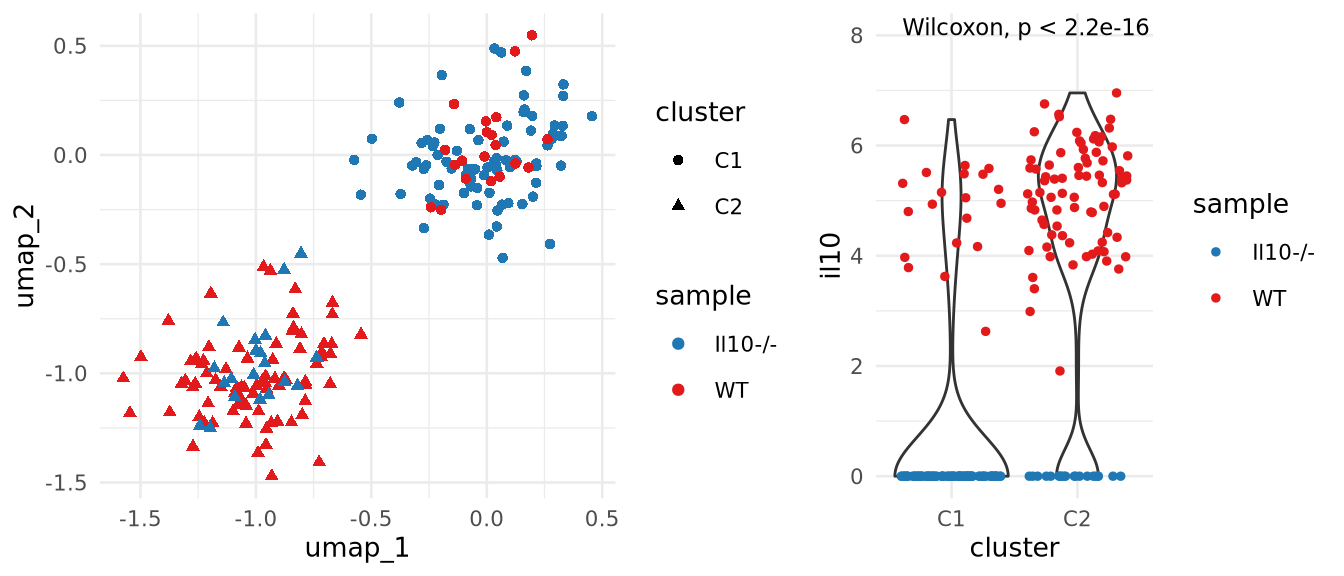
Complex designs: Approaches
Complex designs: Approaches
| Approach | Speed | Can include covariates | Can account for multilevel design | Sensitivity | Specificity |
|---|---|---|---|---|---|
| Naive model | Fast | ✖ | ✖ | High | Low |
| GLM | Slow | ✓ | Not recommended | High | Low |
| Mixed models | Slow | ✓ | ✓ | Medium | Medium |
| Pseudobulk | Fast | ✓ | ✓ | Low | High |
NB: This table broadly summarizes each approach, but each approach includes many methods with their own advantages and disadvantages.
Further reading: Soneson & Robinson (2018) Zimmerman et al. (2021) Juntilla et al. (2022) Das et al. (2022)
1-vs-1 and 1-vs-all

- 1-vs-1: C1 vs C2
- 1-vs-all: C1 vs C0 + C2 + C3
1-vs-all analysis
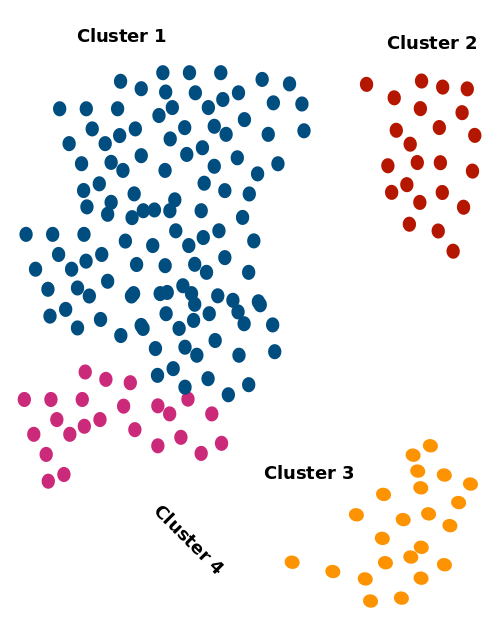
- Larger clusters will be over-represented unless subsampled
- Highly similar clusters
- Will have most of their DEGs overlapping
- Pairwise comparisons might help rather than 1 vs rest
Other considerations
Considerations - p-values
- \(p\) depends on \(n\), variance and intergroup difference
- As \(n\) increases, variance can increase and difference can decrease without losing power
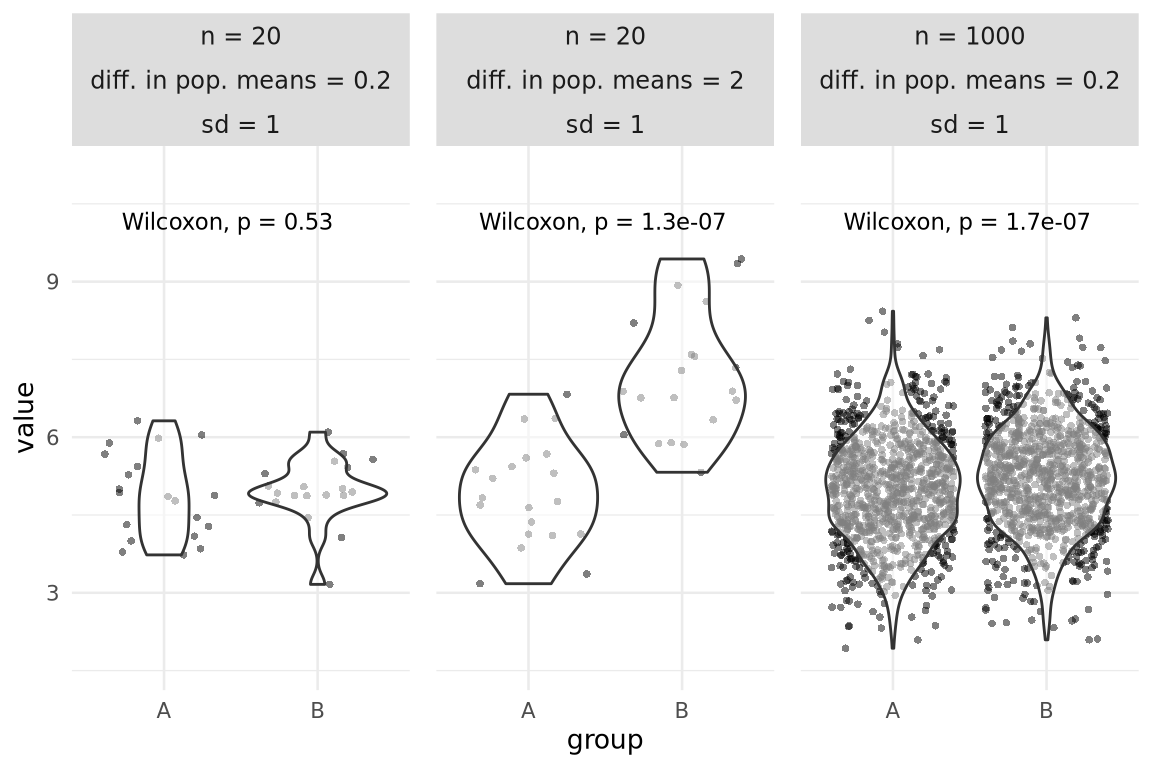
Are all statistically significant differences of interest for your research question?
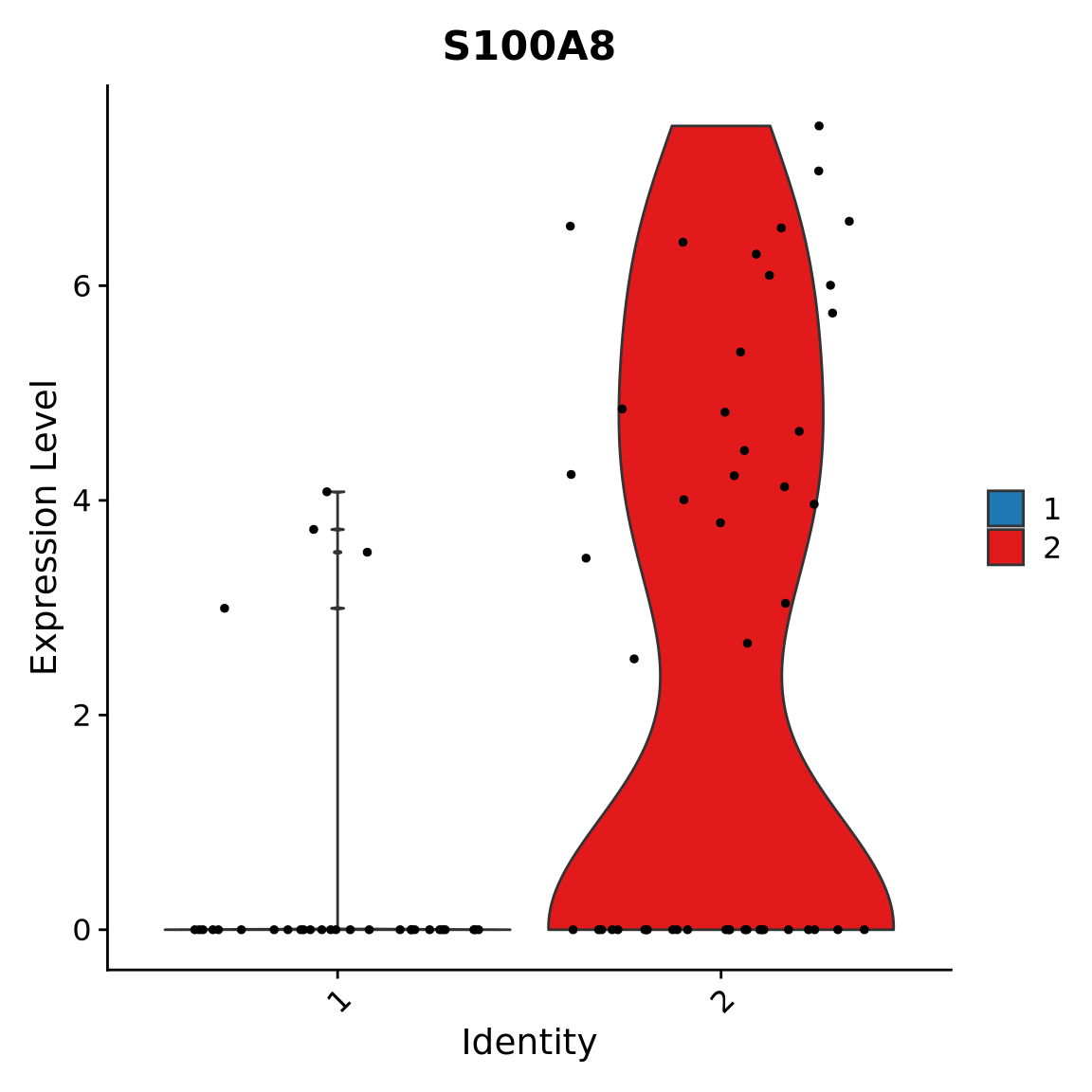
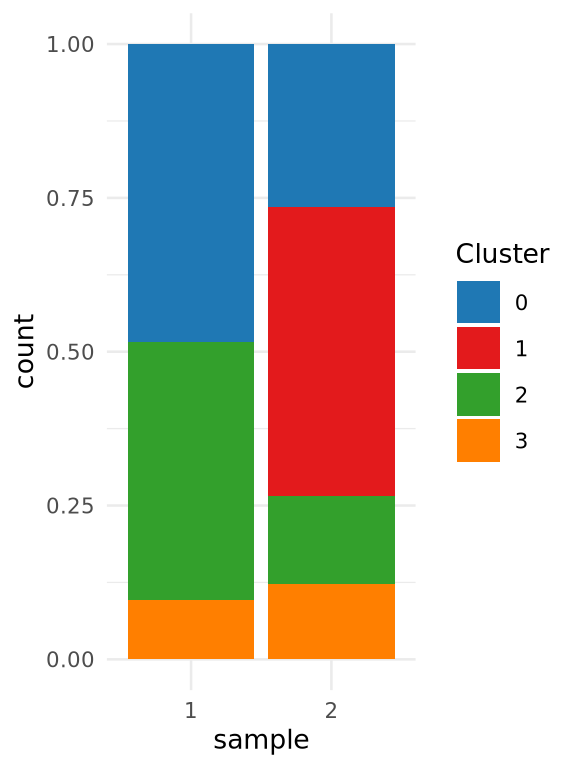
Considerations: Composition vs expression
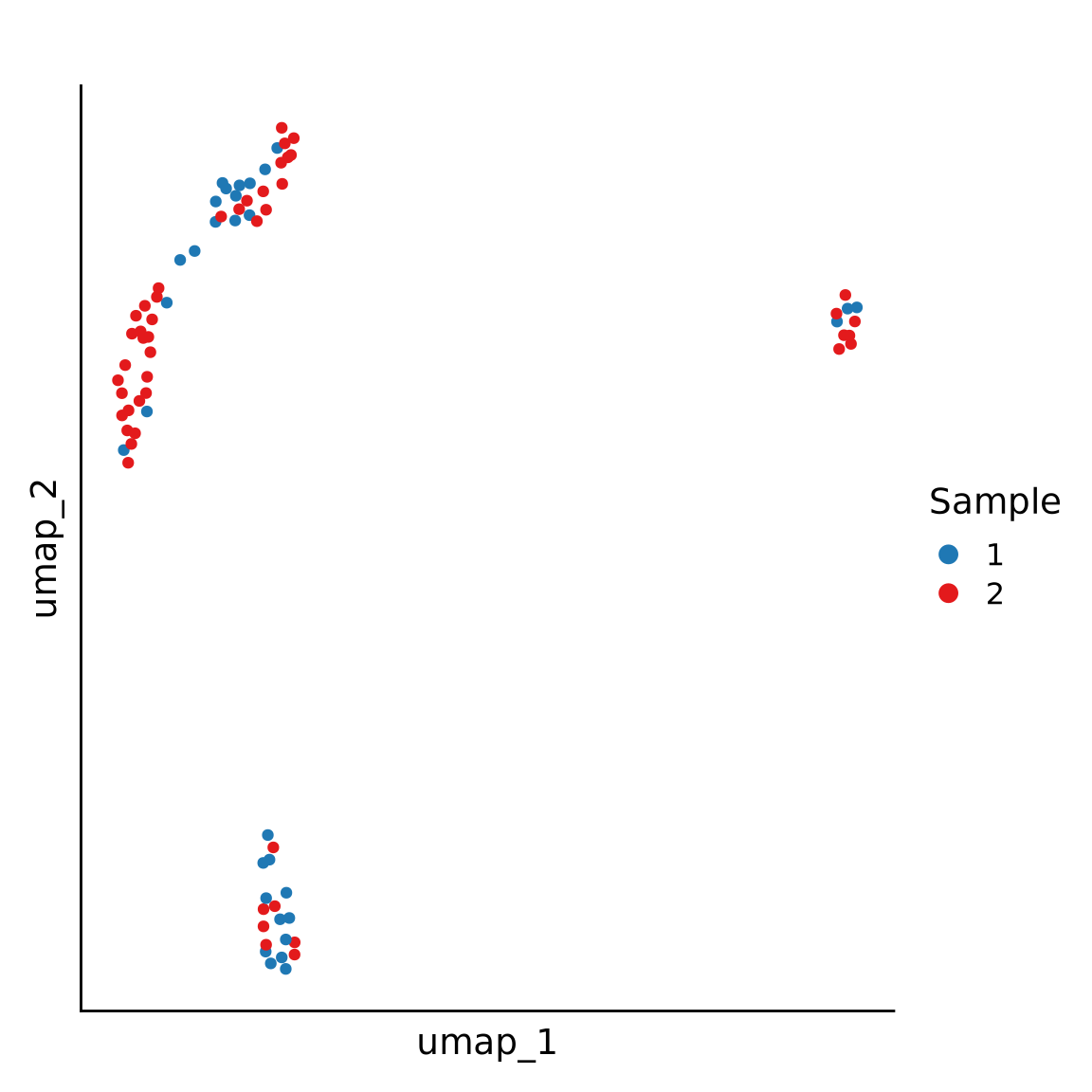
| avg_log2FC | p_val_adj | pct.1 | pct.2 | |
|---|---|---|---|---|
| S100A8 | 5.144138 | 0.0507286 | 0.510 | 0.129 |
| SAT1 | 1.210656 | 0.0858772 | 0.857 | 0.484 |
| TALDO1 | 1.868835 | 0.1229653 | 0.673 | 0.226 |
| GZMM | -2.129484 | 0.1467967 | 0.102 | 0.452 |
| SPON2 | -2.597993 | 0.2331345 | 0.082 | 0.387 |
Assessing results
- Methods are hard to evaluate - we don’t know the ground truth
- Using known data (positive controls)
- Simulated data by modelling
- Intersect of multiple methods
- Visual inspection
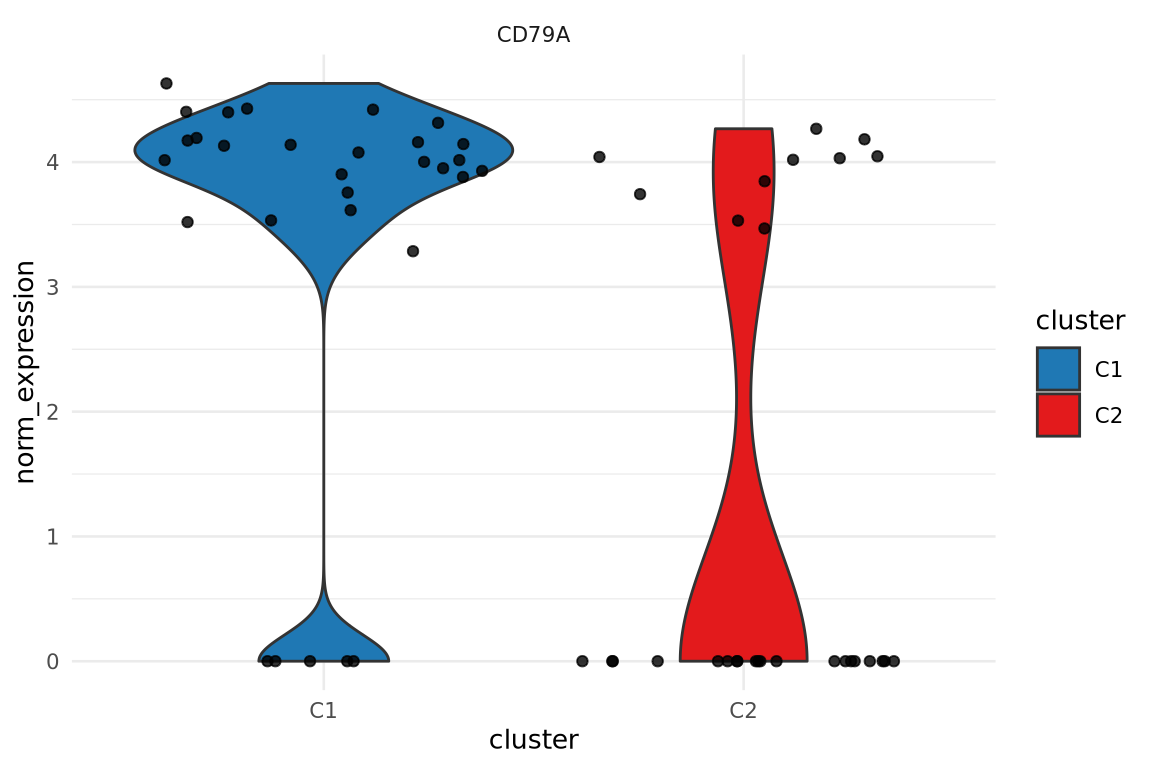
Assessing results
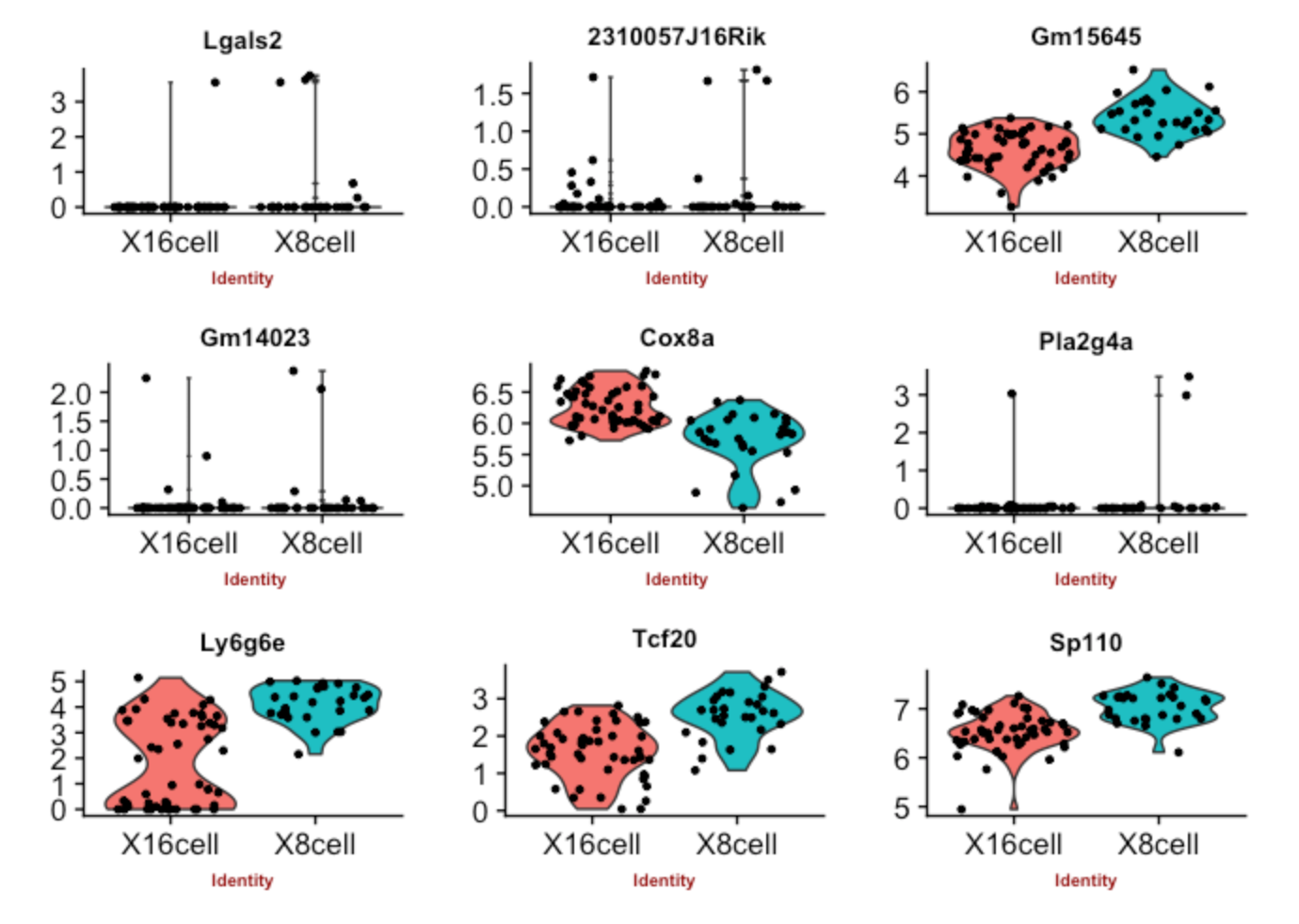
Violin plots are good to visualize distribution
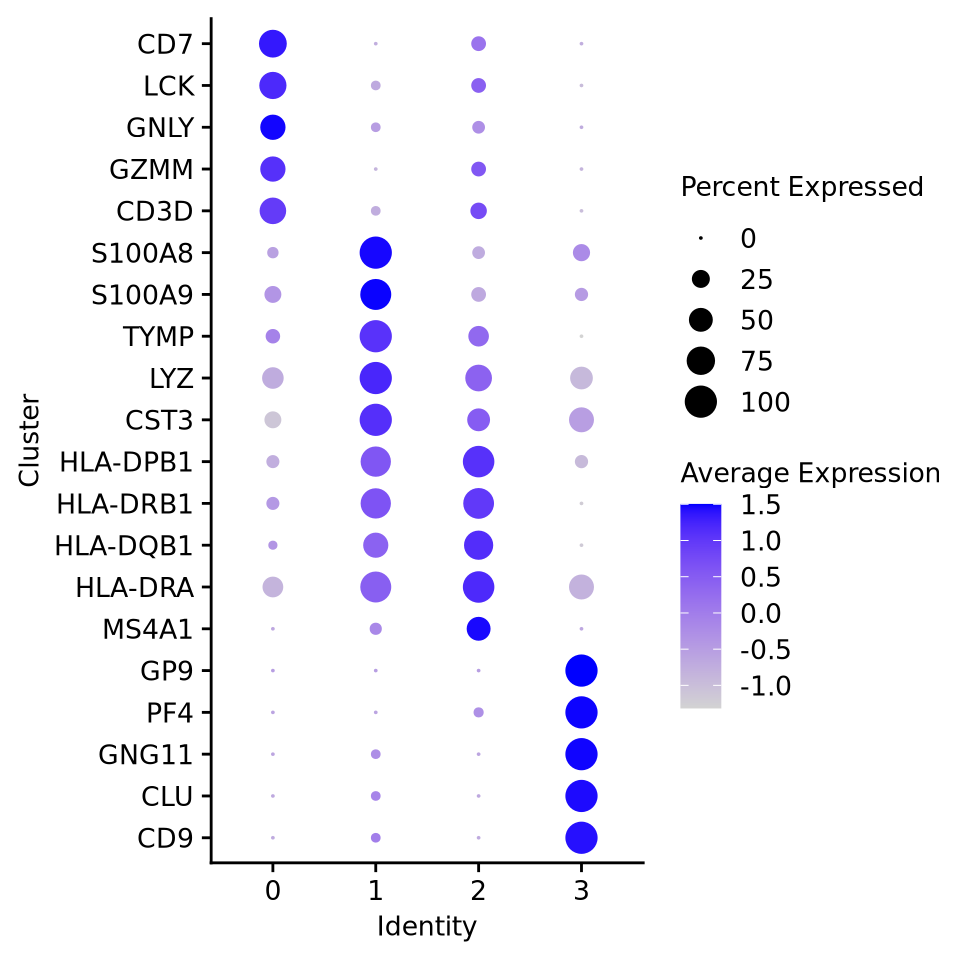
Dot plots give a quick overview of both expression and % of cells expressing a gene
Things to think about
- How many cells/samples do I need for reliable DGE?
- How different do I expect my cells/samples to be?
- How high is the expression and how deep am I sequencing?
- Will also depend on library quality
- Which test should I use?
- Which populations am I comparing?
- Are cells independent within my groups of interest?
- Do I need to correct for any batch effects?
- Which data should I use? Raw? Normalized? Log Normalized?
- Depends on test/method
- DE results are always relative to other cells
- Don’t just rely on p-values
- Always assess your results!
- Visualize the full distributions
- Check for potential confounders
- Batch effects can be corrected using latent variables (assuming good experimental design)
- Removing ambient RNA can also help
Conclusions
- Single cell data is more complex than differences in mean expression
- Different tests rely on different assumptions
- Always consider what you are trying to compare
- Important to assess and validate the results
References
Acknowledgements
Slides adapted from previous presentations by Olga Dethlefson, Åsa Björklund, Vincent van Hoef and Roy Francis.

