# download pre-computed annotation
fetch_annotation <- TRUE
# url for source and intermediate data
path_data <- "https://nextcloud.dc.scilifelab.se/public.php/webdav"
curl_upass <- "-u zbC5fr2LbEZ9rSE:scRNAseq2025"
path_covid <- "./data/covid/raw"
if (!dir.exists(path_covid)) dir.create(path_covid, recursive = T)
path_results <- "./data/covid/results"
if (!dir.exists(path_results)) dir.create(path_results, recursive = T)Code chunks run R commands unless otherwise specified.
1 Get data
In this tutorial, we will run all tutorials with a set of 8 PBMC 10x datasets from 4 covid-19 patients and 4 healthy controls, the samples have been subsampled to 1500 cells per sample. We can start by defining our paths.
file_list <- c(
"normal_pbmc_13.h5", "normal_pbmc_14.h5", "normal_pbmc_19.h5", "normal_pbmc_5.h5",
"ncov_pbmc_15.h5", "ncov_pbmc_16.h5", "ncov_pbmc_17.h5", "ncov_pbmc_1.h5"
)
for (i in file_list) {
path_file <- file.path(path_covid, i)
if (!file.exists(path_file)) {
download.file(url = file.path(file.path(path_data, "covid/raw"), i),
destfile = path_file, method = "curl", extra = curl_upass)
}
}With data in place, now we can start loading libraries we will use in this tutorial.
suppressPackageStartupMessages({
library(scater)
library(scran)
library(patchwork) # combining figures
library(org.Hs.eg.db)
library(scDblFinder)
})We can first load the data individually by reading directly from HDF5 file format (.h5).
cov.15 <- Seurat::Read10X_h5(
filename = file.path(path_covid, "ncov_pbmc_15.h5"),
use.names = T
)
cov.1 <- Seurat::Read10X_h5(
filename = file.path(path_covid, "ncov_pbmc_1.h5"),
use.names = T
)
cov.16 <- Seurat::Read10X_h5(
filename = file.path(path_covid, "ncov_pbmc_16.h5"),
use.names = T
)
cov.17 <- Seurat::Read10X_h5(
filename = file.path(path_covid, "ncov_pbmc_17.h5"),
use.names = T
)
ctrl.5 <- Seurat::Read10X_h5(
filename = file.path(path_covid, "normal_pbmc_5.h5"),
use.names = T
)
ctrl.13 <- Seurat::Read10X_h5(
filename = file.path(path_covid, "normal_pbmc_13.h5"),
use.names = T
)
ctrl.14 <- Seurat::Read10X_h5(
filename = file.path(path_covid, "normal_pbmc_14.h5"),
use.names = T
)
ctrl.19 <- Seurat::Read10X_h5(
filename = file.path(path_covid, "normal_pbmc_19.h5"),
use.names = T
)2 Collate
We can now merge them objects into a single object. Each analysis workflow (Seurat, Scater, Scanpy, etc) has its own way of storing data. We will add dataset labels as cell.ids just in case you have overlapping barcodes between the datasets. After that we add a column type in the metadata to define covid and ctrl samples.
sce <- SingleCellExperiment(assays = list(counts = cbind(cov.1, cov.15, cov.16, cov.17, ctrl.5, ctrl.13, ctrl.14,ctrl.19)))
dim(sce)[1] 33538 12000# Adding metadata
sce@colData$sample <- unlist(sapply(c("cov.1", "cov.15", "cov.16", "cov.17", "ctrl.5", "ctrl.13", "ctrl.14","ctrl.19"), function(x) rep(x, ncol(get(x)))))
sce@colData$type <- ifelse(grepl("cov", sce@colData$sample), "Covid", "Control")Once you have created the merged Seurat object, the count matrices and individual count matrices and objects are not needed anymore. It is a good idea to remove them and run garbage collect to free up some memory.
# remove all objects that will not be used.
rm(cov.15, cov.1, cov.17, cov.16, ctrl.5, ctrl.13, ctrl.14, ctrl.19)
# run garbage collect to free up memory
gc() used (Mb) gc trigger (Mb) max used (Mb)
Ncells 11938593 637.6 17315374 924.8 16951453 905.4
Vcells 47505418 362.5 119270556 910.0 119024651 908.1Here is how the count matrix and the metadata look like for every cell.
head(counts(sce)[, 1:10])6 x 10 sparse Matrix of class "dgCMatrix"
MIR1302-2HG . . . . . . . . . .
FAM138A . . . . . . . . . .
OR4F5 . . . . . . . . . .
AL627309.1 . . . . . . . . . .
AL627309.3 . . . . . . . . . .
AL627309.2 . . . . . . . . . .head(sce@colData, 10)DataFrame with 10 rows and 2 columns
sample type
<character> <character>
AGGTAGGTCGTTGTTT-1 cov.1 Covid
TAGAGTCGTCCTCCAT-1 cov.1 Covid
CCCTGATAGCGAACTG-1 cov.1 Covid
TCATCATTCCACGTAA-1 cov.1 Covid
ATTTACCCAAGCCTGC-1 cov.1 Covid
GTTGTCCTCTAGAACC-1 cov.1 Covid
CCTCCAACAAGAGATT-1 cov.1 Covid
AATAGAGGTGTGAGCA-1 cov.1 Covid
GGTGGCTAGCGAATGC-1 cov.1 Covid
TCGGGCACAGTGTGGA-1 cov.1 Covid3 Calculate QC
Having the data in a suitable format, we can start calculating some quality metrics. We can for example calculate the percentage of mitochondrial and ribosomal genes per cell and add to the metadata. The proportion of hemoglobin genes can give an indication of red blood cell contamination, but in some tissues it can also be the case that some celltypes have higher content of hemoglobin. This will be helpful to visualize them across different metadata parameters (i.e. datasetID and chemistry version). There are several ways of doing this. The QC metrics are finally added to the metadata table.
Citing from Simple Single Cell workflows (Lun, McCarthy & Marioni, 2017): High proportions are indicative of poor-quality cells (Islam et al. 2014; Ilicic et al. 2016), possibly because of loss of cytoplasmic RNA from perforated cells. The reasoning is that mitochondria are larger than individual transcript molecules and less likely to escape through tears in the cell membrane.
# Mitochondrial genes
mito_genes <- rownames(sce)[grep("^MT-", rownames(sce))]
# Ribosomal genes
ribo_genes <- rownames(sce)[grep("^RP[SL]", rownames(sce))]
# Hemoglobin genes - includes all genes starting with HB except HBP.
hb_genes <- rownames(sce)[grep("^HB[^(P|E|S)]", rownames(sce))]First, let Scran calculate some general qc-stats for genes and cells with the function perCellQCMetrics. It can also calculate proportion of counts for specific gene subsets, so first we need to define which genes are mitochondrial, ribosomal and hemoglobin.
sce <- addPerCellQC(sce, flatten = T, subsets = list(mt = mito_genes, hb = hb_genes, ribo = ribo_genes))
# Way2: Doing it manually
sce@colData$percent_mito <- Matrix::colSums(counts(sce)[mito_genes, ]) / sce@colData$total * 100Now you can see that we have additional data in the metadata slot.
head(colData(sce))DataFrame with 6 rows and 15 columns
sample type sum detected subsets_mt_sum
<character> <character> <numeric> <integer> <numeric>
AGGTAGGTCGTTGTTT-1 cov.1 Covid 1396 656 26
TAGAGTCGTCCTCCAT-1 cov.1 Covid 1613 779 186
CCCTGATAGCGAACTG-1 cov.1 Covid 9482 2036 761
TCATCATTCCACGTAA-1 cov.1 Covid 4357 875 2960
ATTTACCCAAGCCTGC-1 cov.1 Covid 12466 3290 686
GTTGTCCTCTAGAACC-1 cov.1 Covid 5541 1606 707
subsets_mt_detected subsets_mt_percent subsets_hb_sum
<integer> <numeric> <numeric>
AGGTAGGTCGTTGTTT-1 8 1.86246 1
TAGAGTCGTCCTCCAT-1 10 11.53131 0
CCCTGATAGCGAACTG-1 11 8.02573 3
TCATCATTCCACGTAA-1 13 67.93665 2
ATTTACCCAAGCCTGC-1 12 5.50297 1
GTTGTCCTCTAGAACC-1 12 12.75943 3
subsets_hb_detected subsets_hb_percent subsets_ribo_sum
<integer> <numeric> <numeric>
AGGTAGGTCGTTGTTT-1 1 0.07163324 11
TAGAGTCGTCCTCCAT-1 0 0.00000000 96
CCCTGATAGCGAACTG-1 1 0.03163889 4157
TCATCATTCCACGTAA-1 2 0.04590314 99
ATTTACCCAAGCCTGC-1 1 0.00802182 2281
GTTGTCCTCTAGAACC-1 2 0.05414185 1664
subsets_ribo_detected subsets_ribo_percent total
<integer> <numeric> <numeric>
AGGTAGGTCGTTGTTT-1 9 0.787966 1396
TAGAGTCGTCCTCCAT-1 45 5.951643 1613
CCCTGATAGCGAACTG-1 85 43.840962 9482
TCATCATTCCACGTAA-1 52 2.272206 4357
ATTTACCCAAGCCTGC-1 82 18.297770 12466
GTTGTCCTCTAGAACC-1 80 30.030680 5541
percent_mito
<numeric>
AGGTAGGTCGTTGTTT-1 1.86246
TAGAGTCGTCCTCCAT-1 11.53131
CCCTGATAGCGAACTG-1 8.02573
TCATCATTCCACGTAA-1 67.93665
ATTTACCCAAGCCTGC-1 5.50297
GTTGTCCTCTAGAACC-1 12.759434 Plot QC
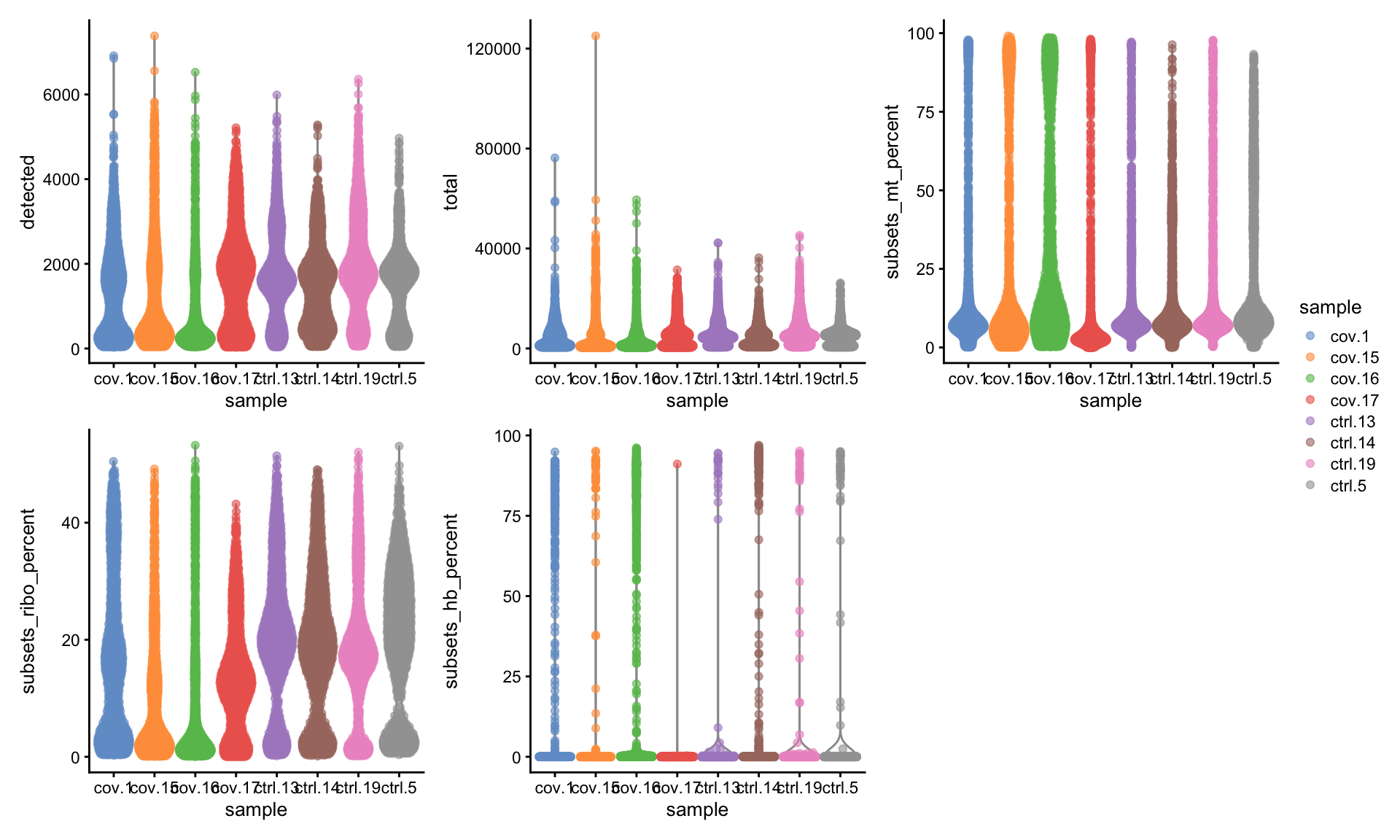
Now we can plot some of the QC variables as violin plots.
# total is total UMIs per cell
# detected is number of detected genes.
# the different gene subset percentages are listed as subsets_mt_percent etc.
wrap_plots(
plotColData(sce, y = "detected", x = "sample", colour_by = "sample"),
plotColData(sce, y = "total", x = "sample", colour_by = "sample"),
plotColData(sce, y = "subsets_mt_percent", x = "sample", colour_by = "sample"),
plotColData(sce, y = "subsets_ribo_percent", x = "sample", colour_by = "sample"),
plotColData(sce, y = "subsets_hb_percent", x = "sample", colour_by = "sample"),
ncol = 3
) + plot_layout(guides = "collect")Looking at the violin plots, what do you think are appropriate cutoffs for filtering these samples
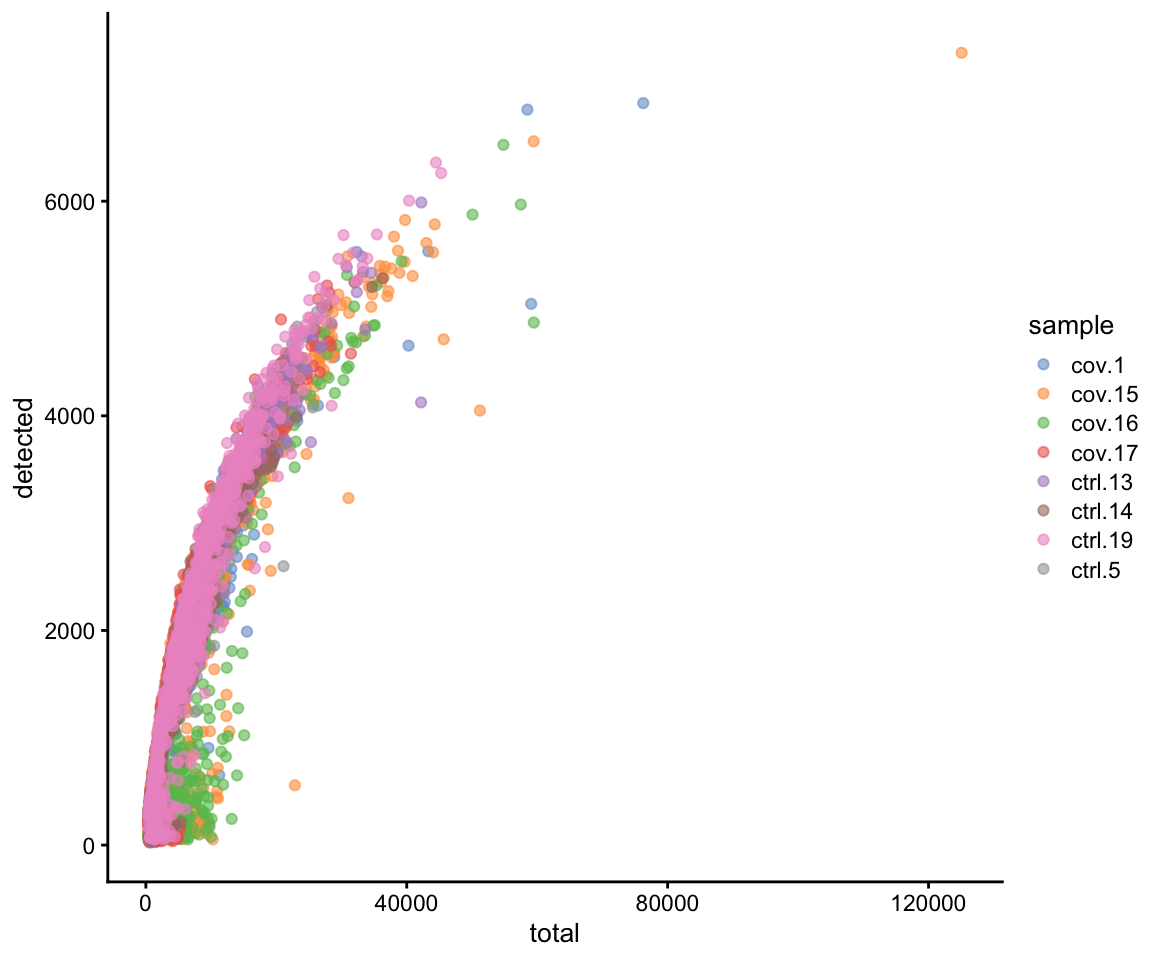
As you can see, there is quite some difference in quality for these samples, with for instance the covid_15 and covid_16 samples having cells with fewer detected genes and more mitochondrial content. As the ribosomal proteins are highly expressed they will make up a larger proportion of the transcriptional landscape when fewer of the lowly expressed genes are detected. We can also plot the different QC-measures as scatter plots.
Plot additional QC stats that we have calculated as scatter plots. How are the different measures correlated? Can you explain why?
5 Filtering
5.1 Detection-based filtering
A standard approach is to filter cells with low number of reads as well as genes that are present in at least a given number of cells. Here we will only consider cells with at least 200 detected genes and genes need to be expressed in at least 3 cells. Please note that those values are highly dependent on the library preparation method used.
In Scran, we can use the function quickPerCellQC to filter out outliers from distributions of qc stats, such as detected genes, gene subsets etc. But in this case, we will take one setting at a time and run through the steps of filtering cells.
dim(sce)[1] 33538 12000selected_c <- colnames(sce)[sce$detected > 200]
selected_f <- rownames(sce)[Matrix::rowSums(counts(sce)) > 3]
sce.filt <- sce[selected_f, selected_c]
dim(sce.filt)[1] 18877 10656Extremely high number of detected genes could indicate doublets. However, depending on the cell type composition in your sample, you may have cells with higher number of genes (and also higher counts) from one cell type. In this case, we will run doublet prediction further down, so we will skip this step now, but the code below is an example of how it can be run:
# skip for now and run doublet detection instead...
# high.det.v3 <- sce.filt$nFeatures > 4100
# high.det.v2 <- (sce.filt$nFeatures > 2000) & (sce.filt$sample_id == "v2.1k")
# remove these cells
# sce.filt <- sce.filt[ , (!high.det.v3) & (!high.det.v2)]
# check number of cells
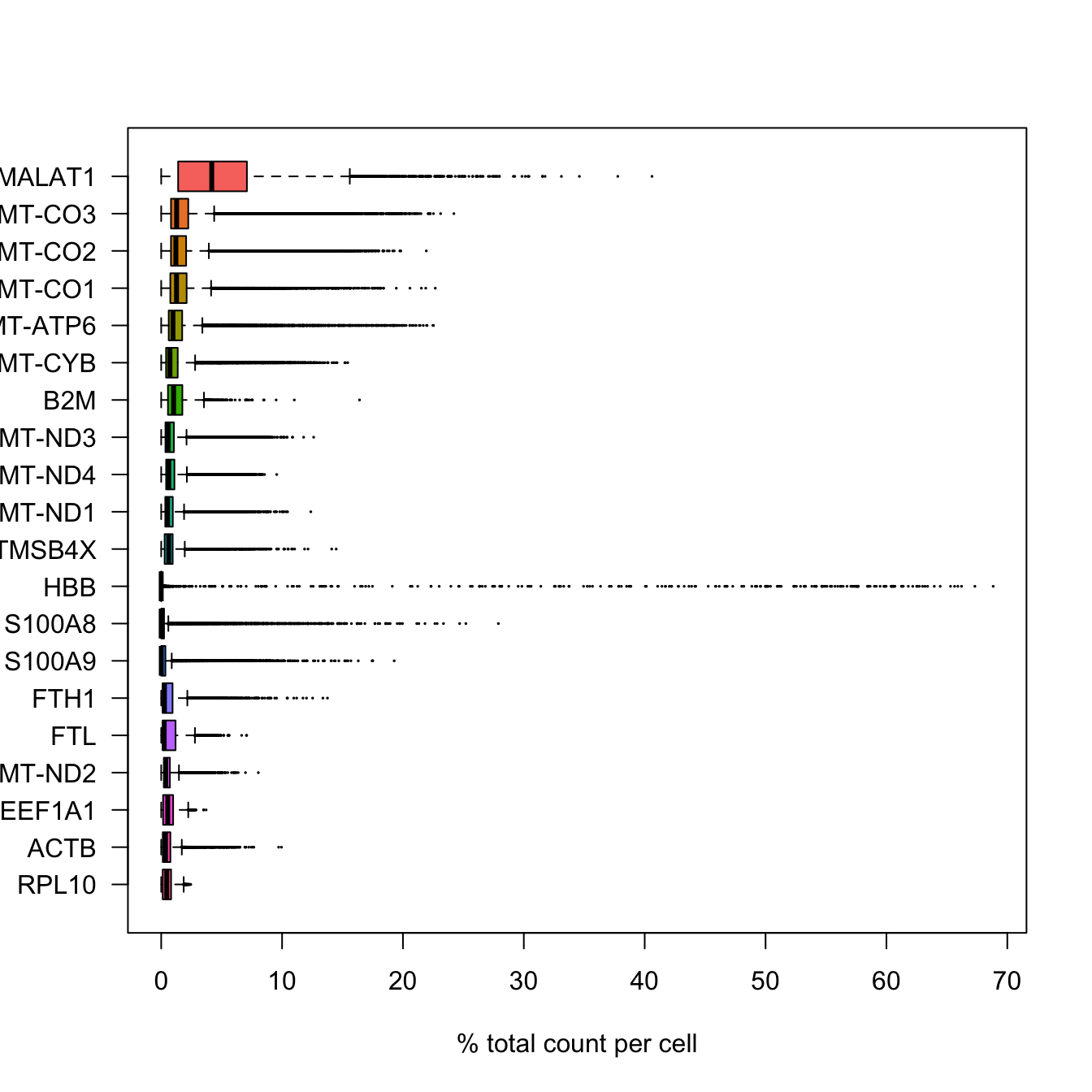
# ncol(sce.filt)Additionally, we can also see which genes contribute the most to such reads. We can for instance plot the percentage of counts per gene.
In Scater, you can also use the function plotHighestExprs() to plot the gene contribution, but the function is quite slow, so we will do it on our own instead..
# Compute the relative expression of each gene per cell
# Use sparse matrix operations, if your dataset is large, doing matrix devisions the regular way will take a very long time.
C <- counts(sce.filt)
C@x <- C@x / rep.int(colSums(C), diff(C@p)) * 100
most_expressed <- order(Matrix::rowSums(C), decreasing = T)[20:1]
boxplot(as.matrix(t(C[most_expressed, ])), cex = .1, las = 1, xlab = "% total count per cell", col = scales::hue_pal()(20)[20:1], horizontal = TRUE)rm(C)
# also, there is the option of running the function "plotHighestExprs" in the scater package, however, this function takes very long to execute.As you can see, MALAT1 constitutes up to 30% of the UMIs from a single cell and the other top genes are mitochondrial and ribosomal genes. It is quite common that nuclear lincRNAs have correlation with quality and mitochondrial reads, so high detection of MALAT1 may be a technical issue. Let us assemble some information about such genes, which are important for quality control and downstream filtering.
5.2 Mito/Ribo filtering
We also have quite a lot of cells with high proportion of mitochondrial and low proportion of ribosomal reads. It would be wise to remove those cells, if we have enough cells left after filtering. Another option would be to either remove all mitochondrial reads from the dataset and hope that the remaining genes still have enough biological signal. A third option would be to just regress out the percent_mito variable during scaling. In this case we had as much as 99.7% mitochondrial reads in some of the cells, so it is quite unlikely that there is much cell type signature left in those. Looking at the plots, make reasonable decisions on where to draw the cutoff. In this case, the bulk of the cells are below 20% mitochondrial reads and that will be used as a cutoff. We will also remove cells with less than 5% ribosomal reads.
selected_mito <- sce.filt$subsets_mt_percent < 20
selected_ribo <- sce.filt$subsets_ribo_percent > 5
# and subset the object to only keep those cells
sce.filt <- sce.filt[, selected_mito & selected_ribo]
dim(sce.filt)[1] 18877 7431As you can see, a large proportion of sample covid_15 is filtered out. Also, there is still quite a lot of variation in percent_mito, so it will have to be dealt with in the data analysis step. We can also notice that the percent_ribo are also highly variable, but that is expected since different cell types have different proportions of ribosomal content, according to their function.
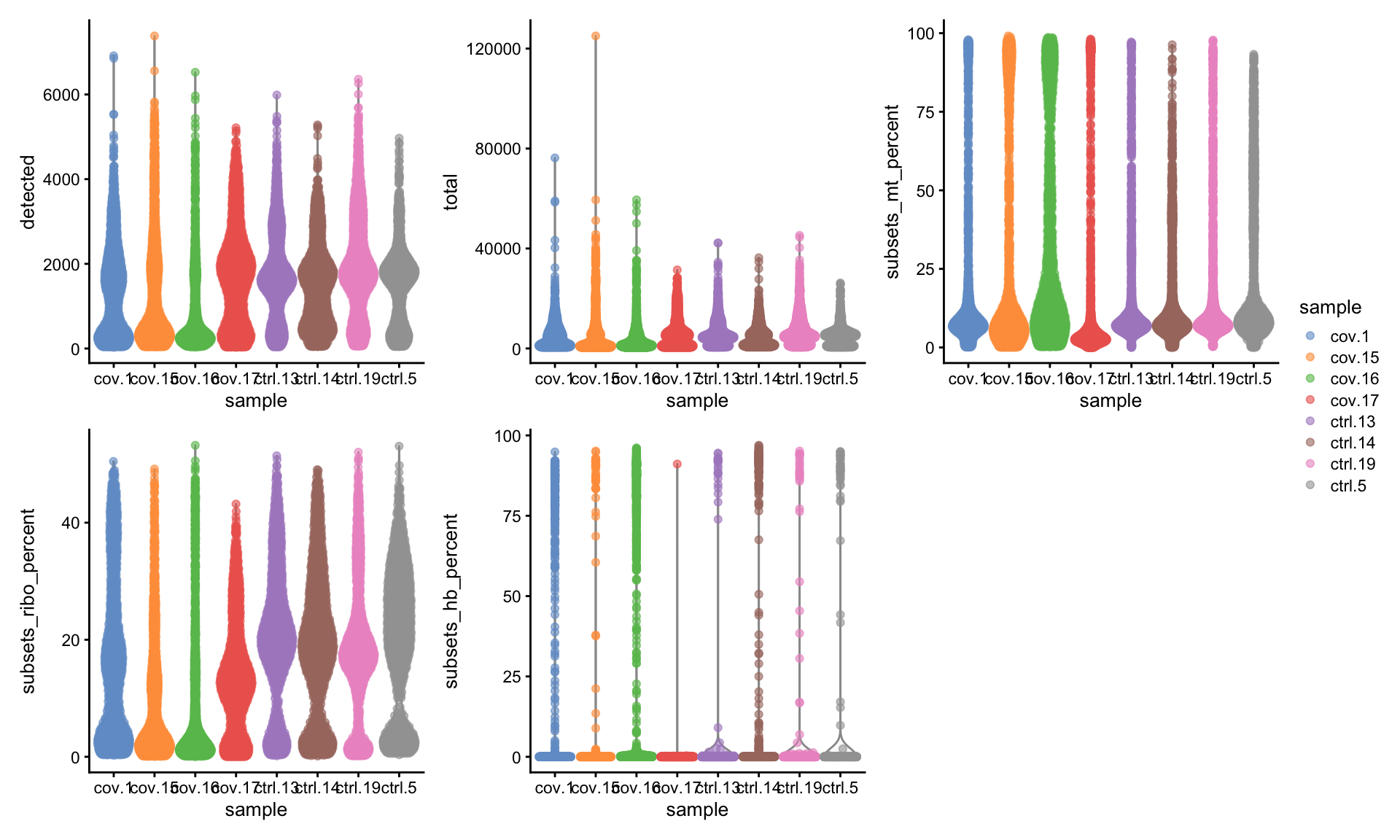
5.3 Plot filtered QC
Lets plot the same QC-stats once more.
wrap_plots(
plotColData(sce, y = "detected", x = "sample", colour_by = "sample"),
plotColData(sce, y = "total", x = "sample", colour_by = "sample"),
plotColData(sce, y = "subsets_mt_percent", x = "sample", colour_by = "sample"),
plotColData(sce, y = "subsets_ribo_percent", x = "sample", colour_by = "sample"),
plotColData(sce, y = "subsets_hb_percent", x = "sample", colour_by = "sample"),
ncol = 3
) + plot_layout(guides = "collect")5.4 Filter genes
As the level of expression of mitochondrial and MALAT1 genes are judged as mainly technical, it can be wise to remove them from the dataset before any further analysis. In this case we will also remove the HB genes.
dim(sce.filt)[1] 18877 7431# Filter MALAT1
sce.filt <- sce.filt[!grepl("MALAT1", rownames(sce.filt)), ]
# Filter Mitocondrial
sce.filt <- sce.filt[!grepl("^MT-", rownames(sce.filt)), ]
# Filter Ribossomal gene (optional if that is a problem on your data)
# sce.filt <- sce.filt[ ! grepl("^RP[SL]", rownames(sce.filt)), ]
# Filter Hemoglobin gene (optional if that is a problem on your data)
sce.filt <- sce.filt[!grepl("^HB[^(P|E|S)]", rownames(sce.filt)), ]
dim(sce.filt)[1] 18854 74316 Sample sex
When working with human or animal samples, you should ideally constrain your experiments to a single sex to avoid including sex bias in the conclusions. However this may not always be possible. By looking at reads from chromosomeY (males) and XIST (X-inactive specific transcript) expression (mainly female) it is quite easy to determine per sample which sex it is. It can also be a good way to detect if there has been any mislabelling in which case, the sample metadata sex does not agree with the computational predictions.
To get chromosome information for all genes, you should ideally parse the information from the gtf file that you used in the mapping pipeline as it has the exact same annotation version/gene naming. However, it may not always be available, as in this case where we have downloaded public data. R package biomaRt can be used to fetch annotation information. The code to run biomaRt is provided. As the biomart instances are quite often unresponsive, we will download and use a file that was created in advance.
Here is the code to download annotation data from Ensembl using biomaRt. We will not run this now and instead use a pre-computed file in the step below.
# fetch_annotation is defined at the top of this document
if (!fetch_annotation) {
suppressMessages(library(biomaRt))
# initialize connection to mart, may take some time if the sites are unresponsive.
mart <- useMart("ENSEMBL_MART_ENSEMBL", dataset = "hsapiens_gene_ensembl")
# fetch chromosome info plus some other annotations
genes_table <- try(biomaRt::getBM(attributes = c(
"ensembl_gene_id", "external_gene_name",
"description", "gene_biotype", "chromosome_name", "start_position"
), mart = mart, useCache = F))
write.csv(genes_table, file = "data/covid/results/genes_table.csv")
}Download precomputed data.
# fetch_annotation is defined at the top of this document
if (fetch_annotation) {
genes_file <- file.path(path_results, "genes_table.csv")
if (!file.exists(genes_file)) download.file(file.path(path_data, "covid/results_bioc/genes_table.csv"), destfile = genes_file,
method = "curl", extra = curl_upass)
}genes.table <- read.csv(genes_file)
genes.table <- genes.table[genes.table$external_gene_name %in% rownames(sce.filt), ]Now that we have the chromosome information, we can calculate the proportion of reads that comes from chromosome Y per cell.But first we have to remove all genes in the pseudoautosmal regions of chrY that are: * chromosome:GRCh38:Y:10001 - 2781479 is shared with X: 10001 - 2781479 (PAR1) * chromosome:GRCh38:Y:56887903 - 57217415 is shared with X: 155701383 - 156030895 (PAR2)
par1 = c(10001, 2781479)
par2 = c(56887903, 57217415)
p1.gene = genes.table$external_gene_name[genes.table$start_position > par1[1] & genes.table$start_position < par1[2] & genes.table$chromosome_name == "Y"]
p2.gene = genes.table$external_gene_name[genes.table$start_position > par2[1] & genes.table$start_position < par2[2] & genes.table$chromosome_name == "Y"]
chrY.gene <- genes.table$external_gene_name[genes.table$chromosome_name == "Y"]
chrY.gene = setdiff(chrY.gene, c(p1.gene, p2.gene))
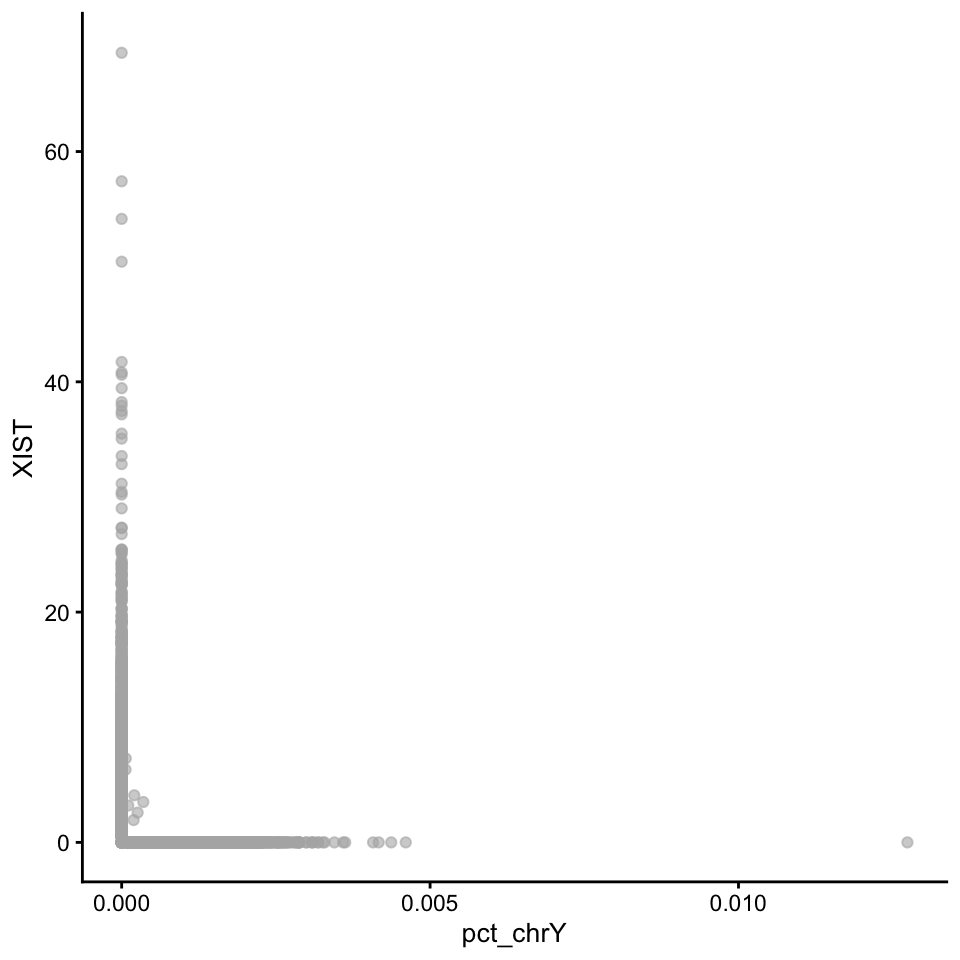
sce.filt@colData$pct_chrY <- Matrix::colSums(counts(sce.filt)[chrY.gene, ]) / colSums(counts(sce.filt))Then plot XIST expression vs chrY proportion. As you can see, the samples are clearly on either side, even if some cells do not have detection of either.
# as plotColData cannot take an expression vs metadata, we need to add in XIST expression to colData
sce.filt@colData$XIST <- counts(sce.filt)["XIST", ] / colSums(counts(sce.filt)) * 10000
plotColData(sce.filt, "XIST", "pct_chrY")Plot as violins.
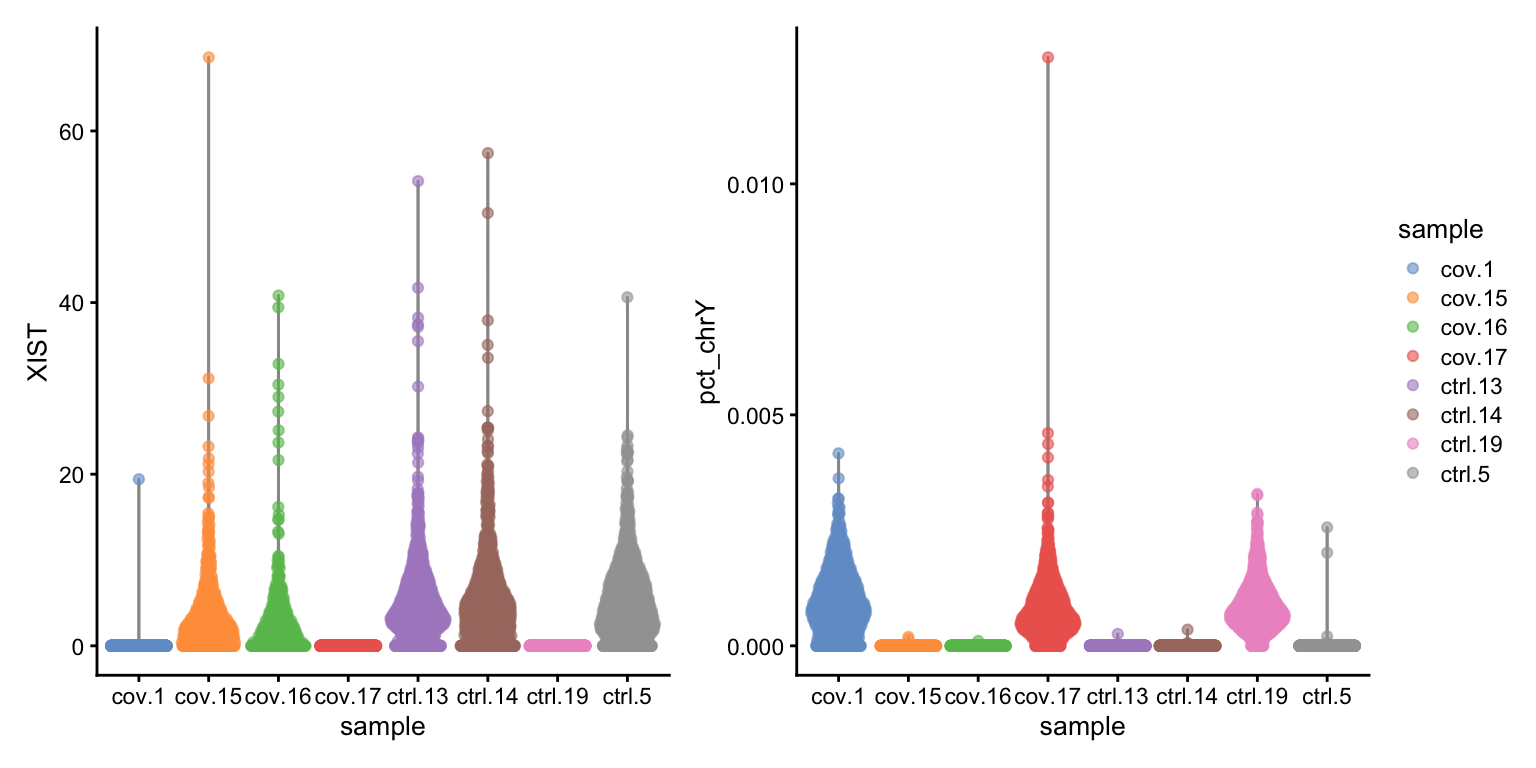
wrap_plots(
plotColData(sce.filt, y = "XIST", x = "sample", colour_by = "sample"),
plotColData(sce.filt, y = "pct_chrY", x = "sample", colour_by = "sample"),
ncol = 2
) + plot_layout(guides = "collect")Here, we can see clearly that we have three males and five females, can you see which samples they are? Do you think this will cause any problems for downstream analysis? Discuss with your group: what would be the best way to deal with this type of sex bias?
7 Cell cycle state
We here perform cell cycle scoring. To score a gene list, the algorithm calculates the difference of mean expression of the given list and the mean expression of reference genes. To build the reference, the function randomly chooses a bunch of genes matching the distribution of the expression of the given list. Cell cycle scoring adds three slots in the metadata, a score for S phase, a score for G2M phase and the predicted cell cycle phase.
hs.pairs <- readRDS(system.file("exdata", "human_cycle_markers.rds", package = "scran"))
anno <- select(org.Hs.eg.db, keys = rownames(sce.filt), keytype = "SYMBOL", column = "ENSEMBL")
ensembl <- anno$ENSEMBL[match(rownames(sce.filt), anno$SYMBOL)]
# Use only genes related to biological process cell cycle to speed up
# https://www.ebi.ac.uk/QuickGO/term/GO:0007049 = cell cycle (BP,Biological Process)
GOs <- na.omit(select(org.Hs.eg.db, keys = na.omit(ensembl), keytype = "ENSEMBL", column = "GO"))
GOs <- GOs[GOs$GO == "GO:0007049", "ENSEMBL"]
hs.pairs <- lapply(hs.pairs, function(x) {
x[rowSums(apply(x, 2, function(i) i %in% GOs)) >= 1, ]
})
str(hs.pairs)List of 3
$ G1 :'data.frame': 6633 obs. of 2 variables:
..$ first : chr [1:6633] "ENSG00000100519" "ENSG00000100519" "ENSG00000100519" "ENSG00000100519" ...
..$ second: chr [1:6633] "ENSG00000065135" "ENSG00000080345" "ENSG00000101266" "ENSG00000135679" ...
$ S :'data.frame': 8308 obs. of 2 variables:
..$ first : chr [1:8308] "ENSG00000255302" "ENSG00000119969" "ENSG00000179051" "ENSG00000127586" ...
..$ second: chr [1:8308] "ENSG00000100519" "ENSG00000100519" "ENSG00000100519" "ENSG00000136856" ...
$ G2M:'data.frame': 6235 obs. of 2 variables:
..$ first : chr [1:6235] "ENSG00000100519" "ENSG00000136856" "ENSG00000136856" "ENSG00000136856" ...
..$ second: chr [1:6235] "ENSG00000146457" "ENSG00000138028" "ENSG00000227268" "ENSG00000101265" ...cc.ensembl <- ensembl[ensembl %in% GOs] # This is the fastest (less genes), but less accurate too
# cc.ensembl <- ensembl[ ensembl %in% unique(unlist(hs.pairs))]
assignments <- cyclone(sce.filt[ensembl %in% cc.ensembl, ], hs.pairs, gene.names = ensembl[ensembl %in% cc.ensembl])
sce.filt$G1.score <- assignments$scores$G1
sce.filt$G2M.score <- assignments$scores$G2M
sce.filt$S.score <- assignments$scores$S
sce.filt$phase <- assignments$phases
table(sce.filt$phase)
G1 G2M S
4322 840 1749 We can now create a violin plot for the cell cycle scores as well.
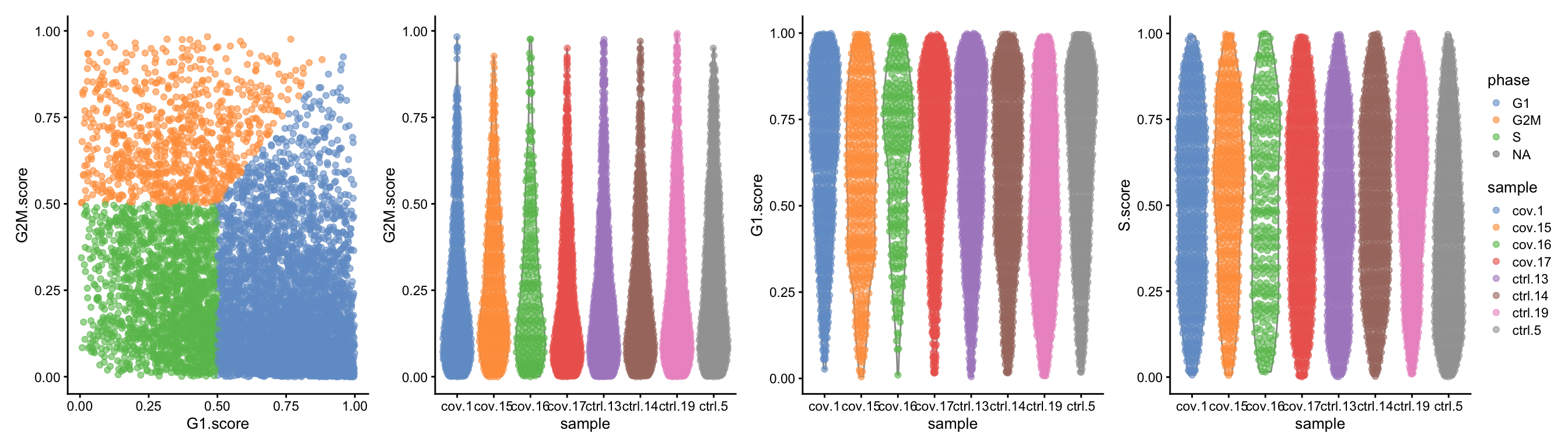
wrap_plots(
plotColData(sce.filt, y = "G2M.score", x = "G1.score", colour_by = "phase"),
plotColData(sce.filt, y = "G2M.score", x = "sample", colour_by = "sample"),
plotColData(sce.filt, y = "G1.score", x = "sample", colour_by = "sample"),
plotColData(sce.filt, y = "S.score", x = "sample", colour_by = "sample"),
ncol = 4
) + plot_layout(guides = "collect")Cyclone predicts most cells as G1, but also quite a lot of cells with high S-Phase scores. Compare to results with Seurat and Scanpy and see how different predictors will give clearly different results.
Cyclone does an automatic prediction of cell cycle phase with a default cutoff of the scores at 0.5 As you can see this does not fit this data very well, so be cautious with using these predictions. Instead we suggest that you look at the scores.
8 Predict doublets
Doublets/Multiples of cells in the same well/droplet is a common issue in scRNAseq protocols. Especially in droplet-based methods with overloading of cells. In a typical 10x experiment the proportion of doublets is linearly dependent on the amount of loaded cells. As indicated from the Chromium user guide, doublet rates are about as follows:
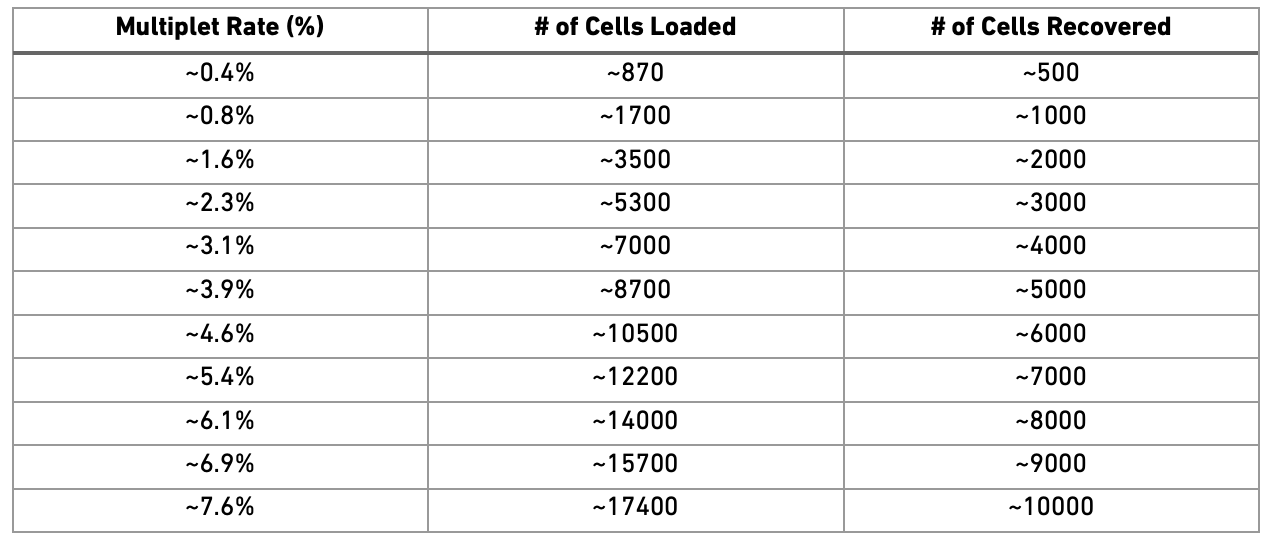
Most doublet detectors simulates doublets by merging cell counts and predicts doublets as cells that have similar embeddings as the simulated doublets. Most such packages need an assumption about the number/proportion of expected doublets in the dataset. The data you are using is subsampled, but the original datasets contained about 5 000 cells per sample, hence we can assume that they loaded about 9 000 cells and should have a doublet rate at about 4%.
Ideally doublet prediction should be run on each sample separately, especially if your samples have different proportions of cell types. In this case, the data is subsampled so we have very few cells per sample and all samples are sorted PBMCs, so it is okay to run them together.
There is a method to predict if a cluster consists of mainly doublets findDoubletClusters(), but we can also predict individual cells based on simulations using the function computeDoubletDensity() which we will do here. Doublet detection will be performed using PCA, so we need to first normalize the data and run variable gene detection, as well as UMAP for visualization. These steps will be explored in more detail in coming exercises.
sce.filt <- logNormCounts(sce.filt)
dec <- modelGeneVar(sce.filt, block = sce.filt$sample)
hvgs <- getTopHVGs(dec, n = 2000)
sce.filt <- runPCA(sce.filt, subset_row = hvgs)
sce.filt <- runUMAP(sce.filt, pca = 10)# run computeDoubletDensity with 10 principal components.
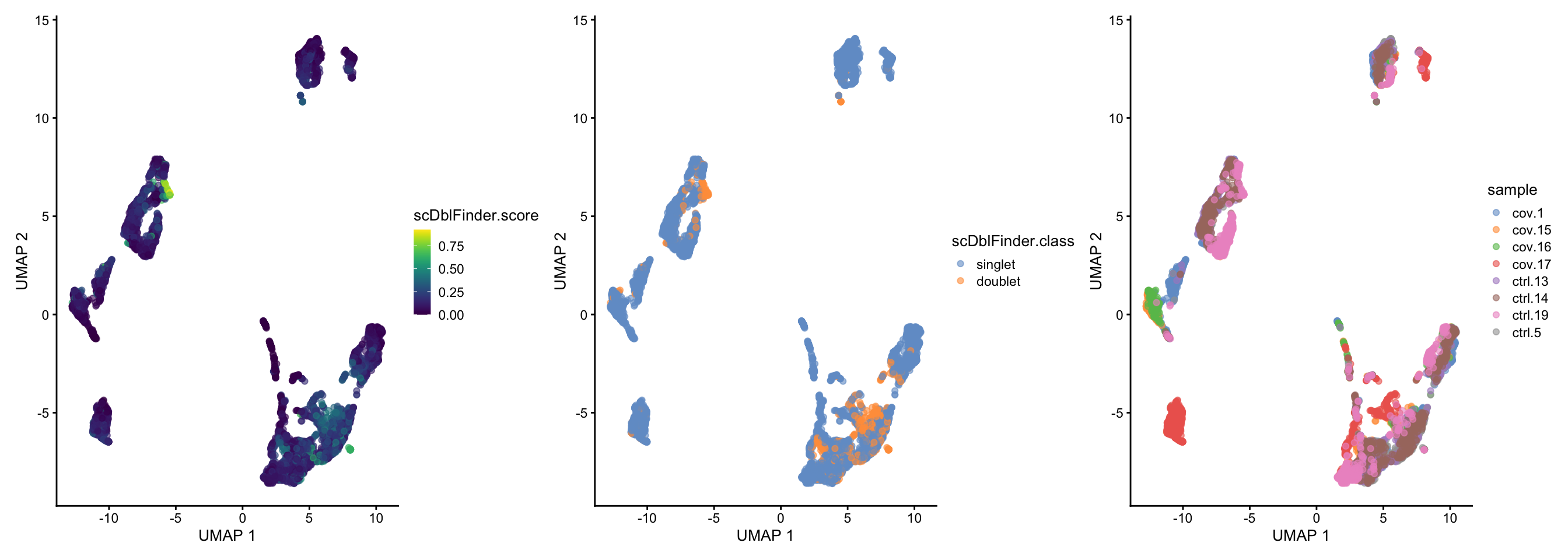
sce.filt <- scDblFinder(sce.filt, dims = 10)wrap_plots(
plotUMAP(sce.filt, colour_by = "scDblFinder.score"),
plotUMAP(sce.filt, colour_by = "scDblFinder.class"),
plotUMAP(sce.filt, colour_by = "sample"),
ncol = 3
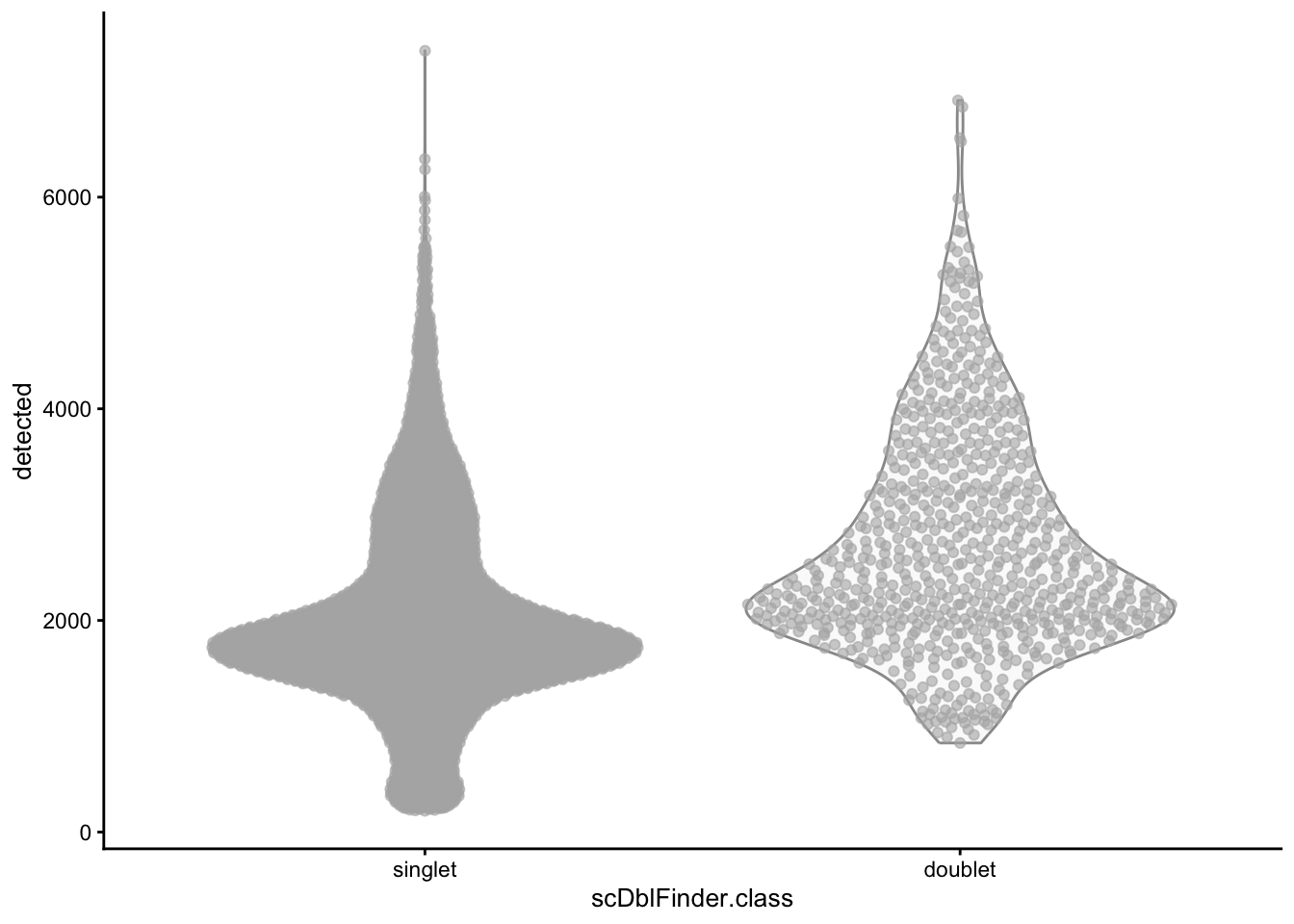
)We should expect that two cells have more detected genes than a single cell, lets check if our predicted doublets also have more detected genes in general.
Now, lets remove all predicted doublets from our data.
sce.filt <- sce.filt[, sce.filt$scDblFinder.class == "singlet"]
dim(sce.filt)[1] 18854 6727To summarize, lets check how many cells we have removed per sample, we started with 1500 cells per sample. Looking back at the intitial QC plots does it make sense that some samples have much fewer cells now?
table(sce$sample)
cov.1 cov.15 cov.16 cov.17 ctrl.13 ctrl.14 ctrl.19 ctrl.5
1500 1500 1500 1500 1500 1500 1500 1500 table(sce.filt$sample)
cov.1 cov.15 cov.16 cov.17 ctrl.13 ctrl.14 ctrl.19 ctrl.5
805 522 348 1018 1103 913 1070 948 “In this case we ran doublet detection with all samples together since we have very small subsampled datasets. But in a real scenario it should be run one sample at a time. Why is this important do you think?”
9 Save data
Finally, lets save the QC-filtered data for further analysis. Create output directory data/covid/results and save data to that folder. This will be used in downstream labs.
saveRDS(sce.filt, file.path(path_results, "bioc_covid_qc.rds"))10 Session info
Click here
sessionInfo()R version 4.3.3 (2024-02-29)
Platform: x86_64-conda-linux-gnu (64-bit)
Running under: Ubuntu 20.04.6 LTS
Matrix products: default
BLAS/LAPACK: /usr/local/conda/envs/seurat/lib/libopenblasp-r0.3.28.so; LAPACK version 3.12.0
locale:
[1] LC_CTYPE=en_US.UTF-8 LC_NUMERIC=C
[3] LC_TIME=en_US.UTF-8 LC_COLLATE=en_US.UTF-8
[5] LC_MONETARY=en_US.UTF-8 LC_MESSAGES=en_US.UTF-8
[7] LC_PAPER=en_US.UTF-8 LC_NAME=C
[9] LC_ADDRESS=C LC_TELEPHONE=C
[11] LC_MEASUREMENT=en_US.UTF-8 LC_IDENTIFICATION=C
time zone: Etc/UTC
tzcode source: system (glibc)
attached base packages:
[1] stats4 stats graphics grDevices utils datasets methods
[8] base
other attached packages:
[1] scDblFinder_1.16.0 org.Hs.eg.db_3.18.0
[3] AnnotationDbi_1.64.1 patchwork_1.3.0
[5] scran_1.30.0 scater_1.30.1
[7] ggplot2_3.5.1 scuttle_1.12.0
[9] SingleCellExperiment_1.24.0 SummarizedExperiment_1.32.0
[11] Biobase_2.62.0 GenomicRanges_1.54.1
[13] GenomeInfoDb_1.38.1 IRanges_2.36.0
[15] S4Vectors_0.40.2 BiocGenerics_0.48.1
[17] MatrixGenerics_1.14.0 matrixStats_1.5.0
loaded via a namespace (and not attached):
[1] RcppAnnoy_0.0.22 splines_4.3.3
[3] later_1.4.1 BiocIO_1.12.0
[5] bitops_1.0-9 tibble_3.2.1
[7] polyclip_1.10-7 XML_3.99-0.17
[9] fastDummies_1.7.5 lifecycle_1.0.4
[11] edgeR_4.0.16 hdf5r_1.3.11
[13] globals_0.16.3 lattice_0.22-6
[15] MASS_7.3-60.0.1 magrittr_2.0.3
[17] limma_3.58.1 plotly_4.10.4
[19] rmarkdown_2.29 yaml_2.3.10
[21] metapod_1.10.0 httpuv_1.6.15
[23] Seurat_5.1.0 sctransform_0.4.1
[25] spam_2.11-1 spatstat.sparse_3.1-0
[27] sp_2.2-0 reticulate_1.40.0
[29] cowplot_1.1.3 pbapply_1.7-2
[31] DBI_1.2.3 RColorBrewer_1.1-3
[33] abind_1.4-5 zlibbioc_1.48.0
[35] Rtsne_0.17 purrr_1.0.2
[37] RCurl_1.98-1.16 GenomeInfoDbData_1.2.11
[39] ggrepel_0.9.6 irlba_2.3.5.1
[41] spatstat.utils_3.1-2 listenv_0.9.1
[43] goftest_1.2-3 RSpectra_0.16-2
[45] spatstat.random_3.3-2 dqrng_0.3.2
[47] fitdistrplus_1.2-2 parallelly_1.42.0
[49] DelayedMatrixStats_1.24.0 leiden_0.4.3.1
[51] codetools_0.2-20 DelayedArray_0.28.0
[53] tidyselect_1.2.1 farver_2.1.2
[55] ScaledMatrix_1.10.0 viridis_0.6.5
[57] spatstat.explore_3.3-4 GenomicAlignments_1.38.0
[59] jsonlite_1.8.9 BiocNeighbors_1.20.0
[61] progressr_0.15.1 ggridges_0.5.6
[63] survival_3.8-3 tools_4.3.3
[65] ica_1.0-3 Rcpp_1.0.14
[67] glue_1.8.0 gridExtra_2.3
[69] SparseArray_1.2.2 xfun_0.50
[71] dplyr_1.1.4 withr_3.0.2
[73] fastmap_1.2.0 bluster_1.12.0
[75] digest_0.6.37 rsvd_1.0.5
[77] R6_2.6.1 mime_0.12
[79] colorspace_2.1-1 scattermore_1.2
[81] tensor_1.5 spatstat.data_3.1-4
[83] RSQLite_2.3.9 tidyr_1.3.1
[85] generics_0.1.3 data.table_1.16.4
[87] rtracklayer_1.62.0 httr_1.4.7
[89] htmlwidgets_1.6.4 S4Arrays_1.2.0
[91] uwot_0.2.2 pkgconfig_2.0.3
[93] gtable_0.3.6 blob_1.2.4
[95] lmtest_0.9-40 XVector_0.42.0
[97] htmltools_0.5.8.1 dotCall64_1.2
[99] SeuratObject_5.0.2 scales_1.3.0
[101] png_0.1-8 spatstat.univar_3.1-1
[103] knitr_1.49 reshape2_1.4.4
[105] rjson_0.2.23 nlme_3.1-167
[107] cachem_1.1.0 zoo_1.8-12
[109] stringr_1.5.1 KernSmooth_2.23-26
[111] parallel_4.3.3 miniUI_0.1.1.1
[113] vipor_0.4.7 restfulr_0.0.15
[115] pillar_1.10.1 grid_4.3.3
[117] vctrs_0.6.5 RANN_2.6.2
[119] promises_1.3.2 BiocSingular_1.18.0
[121] beachmat_2.18.0 xtable_1.8-4
[123] cluster_2.1.8 beeswarm_0.4.0
[125] evaluate_1.0.3 cli_3.6.4
[127] locfit_1.5-9.11 compiler_4.3.3
[129] Rsamtools_2.18.0 rlang_1.1.5
[131] crayon_1.5.3 future.apply_1.11.3
[133] labeling_0.4.3 plyr_1.8.9
[135] ggbeeswarm_0.7.2 stringi_1.8.4
[137] deldir_2.0-4 viridisLite_0.4.2
[139] BiocParallel_1.36.0 munsell_0.5.1
[141] Biostrings_2.70.1 lazyeval_0.2.2
[143] spatstat.geom_3.3-5 Matrix_1.6-5
[145] RcppHNSW_0.6.0 sparseMatrixStats_1.14.0
[147] bit64_4.5.2 future_1.34.0
[149] KEGGREST_1.42.0 statmod_1.5.0
[151] shiny_1.10.0 ROCR_1.0-11
[153] igraph_2.0.3 memoise_2.0.1
[155] bit_4.5.0.1 xgboost_2.1.4.1