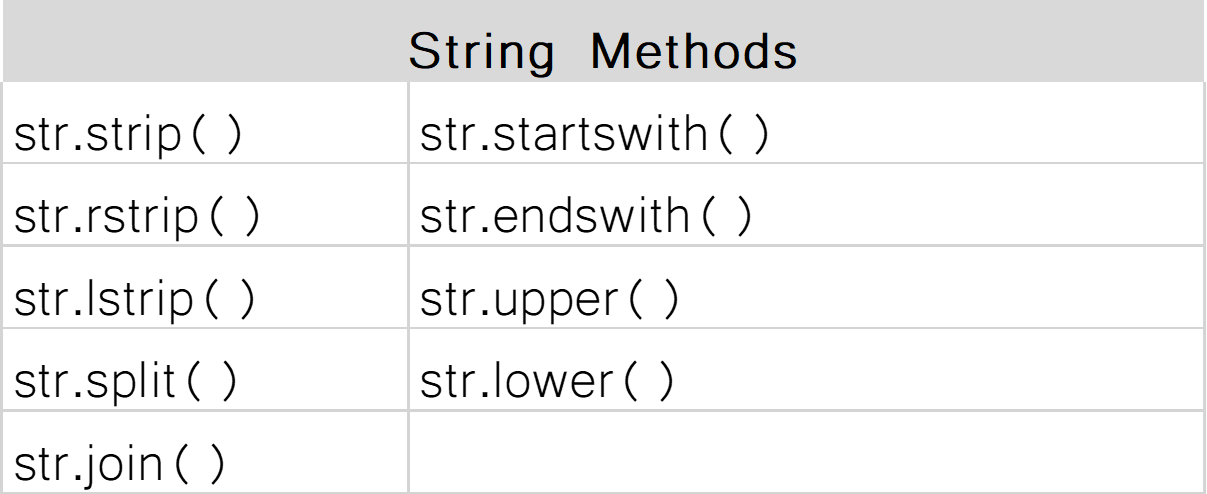
Review Day 1¶
Give an example of the following:
- A number of type
float - A variable containing an
integer - A
Boolean/ Alist/ Astring - What character represents a comment?
- What happens if I take a
listplus alist? - How do I find out if x is present in a
list? - How do I find out if 5 is larger than 3 and the integer 4 is the same as the float 4?
- How do I find the second item in a
list? - An example of a
mutable sequence - An example of an
immutable sequence - Something
iterable(apart from a list) - How do I do to print ‘Yes’ if x is bigger than y?
- How do I open a file handle to read a file called ‘somerandomfile.txt’?
- The file contains several lines, how do I print each line?
Variables and Types¶
A number of type float:
3.14
A variable containing an integer:
a = 5
x = 349852
A boolean:
True
A list:
[2,6,4,8,9]
A string:
'this is a string'
Literals¶
All literals have a type:
- Strings (str) ‘Hello’ “Hi”
- Integers (int) 5
- Floats (float) 3.14
- Boolean (bool) True or False
type(5)
int
Variables¶
Used to store values and to assign them a name.
a = 3.14
a
3.14
Lists¶
A collection of values.
x = [1,5,3,7,8]
y = ['a','b','c']
type(x)
list
Operations¶
What character represents a comment?
#
What happens if I take a list plus a list?
The lists will be concatenated
How do I find out if x is present in a list?
x in [1,2,3,4]
How do I find out if 5 is larger than 3 and the integer 4 is the same as the float 4?
5 > 3 and 4 == 4.0
a = 2
b = 5.46
c = [1,2,3,4]
d = [5,6,7,8]
e = 7
e+a
9
Comparison/Logical/Membership operators¶
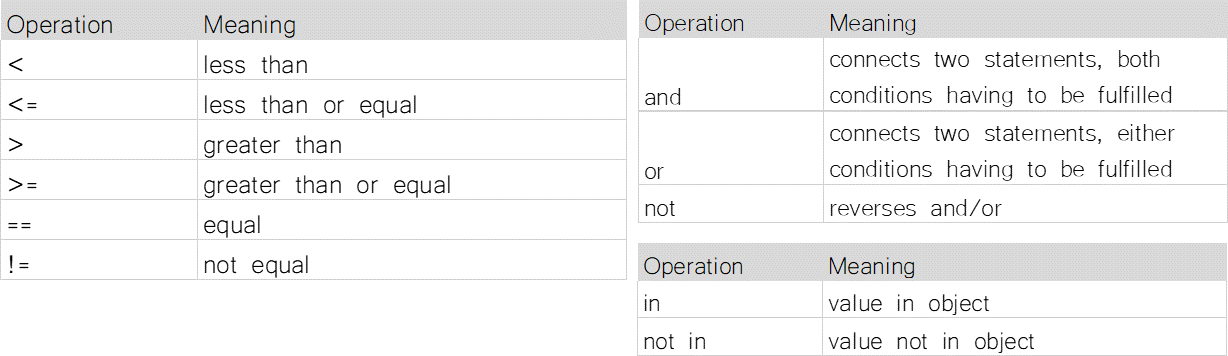
a = [1,2,3,4,5,6,7,8]
b = 5
c = 10
b not in a
False
Sequences¶
How do I find the second item in a list?
list_a[1]
An example of a mutable sequence:
[1,2,3,4,5,6]
An example of an immutable sequence:
'a string is immutable'
Something iterable (apart from a list):
'a string is also iterable'
Indexing¶
Lists (and strings) are an ORDERED collection of elements where every element can be access through an index.
a[0] : first item in list a
REMEMBER! Indexing starts at 0 in python
a = [1,2,3,4,5]
b = ['a','b','c']
c = 'a random string'
a[::2]
a[0:6:2]
[1, 3, 5]
Mutable / Immutable sequences and iterables¶
Lists are mutable object, meaning you can use an index to change the list, while strings are immutable and therefore not changeable.
An iterable sequence is anything you can loop over, ie, lists and strings.
a = [1,2,3,4,5] # mutable
b = ['a','b','c'] # mutable
c = 'a random string' # immutable
#c[0] = 'A'
a[0] = 42
a
[42, 2, 3, 4, 5]
New data type: tuples¶
- A tuple is an immutable sequence of objects
- Unlike a list, nothing can be changed in a tuple
- Still iterable
myTuple = (1,2,3,4,'a','b','c',[42,43,44])
#myTuple[0] = 42
print(myTuple)
#print(len(myTuple))
for i in myTuple:
print(i)
(1, 2, 3, 4, 'a', 'b', 'c', [42, 43, 44]) 1 2 3 4 a b c [42, 43, 44]
If/ Else statements¶
How do I do if I want to print ‘Yes’ if x is bigger than y?
if x > y:
print('Yes')
a = 2
b = [1,2,3,4]
if a in b:
print(str(a)+' is found in the list b')
else:
print(str(a)+' is not in the list')
2 is found in the list b
Files and loops¶
How do I open a file handle to read a file called ‘somerandomfile.txt’?
fh = open('somerandomfile.txt', 'r', encoding = 'utf-8')
fh.close()
The file contains several lines, how do I print each line?
for line in fh:
print(line.strip())
fh = open('../files/somerandomfile.txt','r', encoding = 'utf-8')
for line in fh:
print(line.strip())
fh.close()
just a strange file with some nonsense lines
numbers = [5,6,7,8]
i = 0
while i < len(numbers):
print(numbers[i])
i += 1
5 6 7 8
Questions?¶
Day 2¶
- Pseudocode
- Functions vs Methods
How to approach a coding task¶
Problem:
You have a VCF file with a larger number of samples. You are interested in only one of the samples (sample1) and one region (chr5, 1.000.000-1.005.000). What you want to know is whether this sample has any variants in this region, and if so, what variants.
Always write pseudocode!¶
Pseudocode is a description of what you want to do without actually using proper syntax
Basic Pseudocode:¶
- Open file and loop over lines (ignore lines with #)
- Identify lines where chromosome is 5 and position is between 1.000.000 and 1.005.000
- Isolate the column that contains the genotype for sample1
- Extract the genotypes only from the column
- Check if the genotype contains any alternate alleles
- Print any variants containing alternate alleles for this sample between specified region

- Open file and loop over lines (ignore lines starting with #)
fh = open('C:/Users/Nina/Documents/courses/Python_Beginner_Course/genotypes.vcf', 'r', encoding = 'utf-8')
for line in fh:
if not line.startswith('#'):
print(line.strip())
break
fh.close()
# Next, find chromosome 5
1 10492 . C T 550.31 LOW_VQSLOD AN=26;AC=2 GT:AD:DP:GQ:PGT:PID:PL ./.:0,0:0:.:.:.:. ./.:0,0:0:.:.:.:. ./.:0,0:0:.:.:.:. ./.:0,0:0:.:.:.:. ./.:0,0:0:.:.:.:. 0/1:12,7:19:99:0|1:10403_ACCCTAACCCTAACCCTAACCCTAACCCTAACCCTAAC_A:196,0,340 ./.:0,0:0:.:.:.:. ./.:0,0:0:.:.:.:. ./.:0,0:0:.:.:.:. ./.:0,0:0:.:.:.:. 0/1:18,4:22:48:.:.:48,0,504 ./.:0,0:0:.:.:.:. ./.:0,0:0:.:.:.:.
- Identify lines where chromosome is 5 and position is between 1.000.000 and 1.005.000

fh = open('C:/Users/Nina/Documents/courses/Python_Beginner_Course/genotypes.vcf', 'r', encoding = 'utf-8')
for line in fh:
if not line.startswith('#'):
cols = line.strip().split('\t')
if cols[0] == '5':
print(cols[0])
break
fh.close()
# Next, find the correct region
5

fh = open('C:/Users/Nina/Documents/courses/Python_Beginner_Course/genotypes.vcf', 'r', encoding = 'utf-8')
for line in fh:
if not line.startswith('#'):
cols = line.strip().split('\t')
if cols[0] == '5' and \
int(cols[1]) >= 1000000 and int(cols[1]) <= 1005000:
print(line)
break
fh.close()
# Next, find the genotypes for sample1
5 1000080 . A T 2557.1 PASS AN=26;AC=2 GT:AD:DP:GQ:PL 0/1:15,18:33:99:489,0,357 ./.:0,0:0:.:. ./.:0,0:0:.:. ./.:0,0:0:.:. ./.:0,0:0:.:. ./.:0,0:0:.:. ./.:0,0:0:.:. ./.:0,0:0:.:. 0/1:21,19:40:99:481,0,542 ./.:0,0:0:.:. ./.:0,0:0:.:. ./.:0,0:0:.:. ./.:0,0:0:.:.
- Isolate the column that contains the genotype for sample1

fh = open('C:/Users/Nina/Documents/courses/Python_Beginner_Course/genotypes.vcf', 'r', encoding = 'utf-8')
for line in fh:
if not line.startswith('#'):
cols = line.strip().split('\t')
if cols[0] == '5' and \
int(cols[1]) >= 1000000 and int(cols[1]) <= 1005000:
geno = cols[9]
print(geno)
break
fh.close()
# Next, extract the genotypes only
0/1:15,18:33:99:489,0,357
- Extract the genotypes only from the column

fh = open('C:/Users/Nina/Documents/courses/Python_Beginner_Course/genotypes.vcf', 'r', encoding = 'utf-8')
for line in fh:
if not line.startswith('#'):
cols = line.strip().split('\t')
if cols[0] == '5' and \
int(cols[1]) >= 1000000 and int(cols[1]) <= 1005000:
geno = cols[9].split(':')[0]
print(geno)
break
fh.close()
# Next, find in which positions sample1 has alternate alleles
0/1
- Check if the genotype contains any alternate alleles

fh = open('C:/Users/Nina/Documents/courses/Python_Beginner_Course/genotypes.vcf', 'r', encoding = 'utf-8')
for line in fh:
if not line.startswith('#'):
cols = line.strip().split('\t')
if cols[0] == '5' and \
int(cols[1]) >= 1000000 and int(cols[1]) <= 1005000:
geno = cols[9].split(':')[0]
if geno in ['0/1', '1/1']:
print(geno)
fh.close()
#Next, print nicely
0/1 0/1 0/1 0/1 0/1 0/1 0/1 0/1 0/1 0/1 0/1 0/1 0/1 0/1 0/1 0/1 0/1 0/1 0/1 0/1 0/1 0/1
- Print any variants containing alternate alleles for this sample between specified region

fh = open('C:/Users/Nina/Documents/courses/Python_Beginner_Course/genotypes.vcf', 'r', encoding = 'utf-8')
for line in fh:
if not line.startswith('#'):
cols = line.strip().split('\t')
if cols[0] == '5' and \
int(cols[1]) >= 1000000 and int(cols[1]) <= 1005000:
geno = cols[9].split(':')[0]
if geno in ['0/1', '1/1']:
var = cols[0]+':'+cols[1]+'_'+cols[3]+'-'+cols[4]
print(var+' has genotype: '+geno)
fh.close()
5:1000080_A-T has genotype: 0/1 5:1000156_G-A has genotype: 0/1 5:1001097_C-A has genotype: 0/1 5:1001193_C-T has genotype: 0/1 5:1001245_T-C has genotype: 0/1 5:1001339_C-T has genotype: 0/1 5:1001344_G-C has genotype: 0/1 5:1001683_G-T has genotype: 0/1 5:1001755_G-A has genotype: 0/1 5:1002374_G-A has genotype: 0/1 5:1002382_G-C has genotype: 0/1 5:1002620_T-C has genotype: 0/1 5:1002722_G-A has genotype: 0/1 5:1002819_C-A has genotype: 0/1 5:1003043_G-T has genotype: 0/1 5:1003099_C-T has genotype: 0/1 5:1003135_G-A has genotype: 0/1 5:1004648_A-G has genotype: 0/1 5:1004650_A-C has genotype: 0/1 5:1004665_A-G has genotype: 0/1 5:1004702_G-T has genotype: 0/1 5:1004879_T-C has genotype: 0/1
→ Notebook Day_2_Exercise_1 (~50 minutes)
Comments for Exercise 1¶
fh = open('../downloads/genotypes_small.vcf', 'r', encoding = 'utf-8')
wt = 0
het = 0
hom = 0
for line in fh:
if not line.startswith('#'):
cols = line.strip().split('\t')
chrom = cols[0]
pos = cols[1]
if chrom == '2' and pos == '136608646':
for geno in cols[9:]:
alleles = geno[0:3]
if alleles == '0/0':
wt += 1
elif alleles == '0/1':
het += 1
elif alleles == '1/1':
hom += 1
freq = (2*hom + het)/((wt+hom+het)*2)
print('The frequency of the rs4988235 SNP is: '+str(freq))
fh.close()
The frequency of the rs4988235 SNP is: 0.7833333333333333
with open('../downloads/genotypes_small.vcf', 'r', encoding = 'utf-8') as fh:
for line in fh:
if line.startswith('2\t136608646'):
alleles = [int(item) for sub in [geno[0:3].split('/') \
for geno in line.strip().split('\t')[9:]] \
for item in sub]
print('The frequency of the rs4988235 SNP is: '\
+str(sum(alleles)/len(alleles)))
break
The frequency of the rs4988235 SNP is: 0.7833333333333333
Although much shorter, but maybe not as intuitive...
with open('../downloads/genotypes_small.vcf', 'r', encoding = 'utf-8') as fh:
for line in fh:
if line.startswith('2\t136608646'):
genoInfo = [geno for geno in line.strip().split('\t')[9:]] # extract comlete geno info to list
genotypes = [g[0:3].split('/') for g in genoInfo] # split into alleles to nested list
alleles = [int(item) for sub in genotypes for item in sub] # flatten the nested list to normal list
print('The frequency of the rs4988235 SNP is: '+str(sum(alleles)/len(alleles))) # use sum and len to calculate freq
break
The frequency of the rs4988235 SNP is: 0.7833333333333333
Shorter than the first version, but easier to follow than the second version
More useful functions and methods¶
What is the difference between a function and a method?
A method always belongs to an object of a specific class, a function does not have to. For example:
print('a string') and print(42) both works, even though one is a string and one is an integer
'a string '.strip() works, but [1,2,3,4].strip() does not work. strip() is a method that only works on strings
What does it matter to me?
For now, you mostly need to be aware of the difference, and know the different syntaxes:
A function:
functionName()
A method:
<object>.methodName()
len([1,2,3])
len('a string')
'a string '.strip()
#[1,2,3].strip()
'a string'
Functions¶


float(3)
3.0

max([1,2,35,23,88,4])
88
sum([1,2,3,4],4)
help(sum)
Help on built-in function sum in module builtins:
sum(iterable, start=0, /)
Return the sum of a 'start' value (default: 0) plus an iterable of numbers
When the iterable is empty, return the start value.
This function is intended specifically for use with numeric values and may
reject non-numeric types.

round(3.234556, 2)
3.23
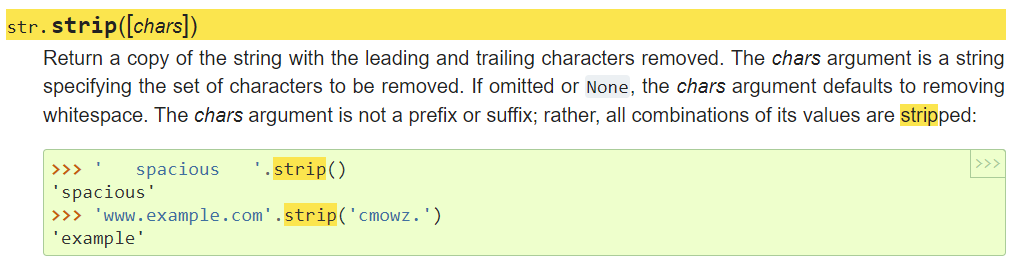

' spaciousWith5678.com'.strip('mco')
' spaciousWith5678.'

a = ' split a string into a list '
a.split(maxsplit=3)
['split', 'a', 'string', 'into a list ']

' '.join('a string already')
#'&'.join(['a', 'b', 'c', 'd'])
'a s t r i n g a l r e a d y'


#'long string'.startswith('ng',2)
'long string'.endswith('string')
True


#'LongRandomString'.lower()
'LongRandomString'.upper()
'LONGRANDOMSTRING'
a = [1,2,3,4,5,5,5,5]
a.append(6)
a.pop()
a.reverse()
a
[5, 5, 5, 5, 4, 3, 2, 1]
Summary¶
- Tuples are immutable sequences of objects
- Always plan your approach before you start coding
- A method always belongs to an object of a specific class, a function does not have to
- The official Python documentation describes the syntax for all built-in functions and methods
→ Notebook Day_2_Exercise_2 (~30 minutes)
IMDb¶
Download the 250.imdb file from the course website
This format of this file is:
- Line by line
- Columns separated by the | character
- Header starting with #

# Votes | Rating | Year | Runtime | URL | Genres | Title
Find the movie with the highest rating¶


fh = open('../downloads/250.imdb', 'r', encoding = 'utf-8')
best = [0,''] # here we save the rating and which movie
for line in fh:
if not line.startswith('#'):
cols = line.strip().split('|')
rating = float(cols[1].strip())
if rating > best[0]: # if the rating is higher than previous highest, update best
best = [rating,cols[6]]
fh.close()
print(best)
[9.3, 'The Shawshank Redemption']
Answer¶
Top movie:
The LOTR: The Return of the King with 8.9
fh = open('../downloads/250.imdb', 'r', encoding = 'utf-8')
top = [0,'']
for line in fh:
if not line.startswith('#'):
cols = line.strip().split('|')
genre = cols[5].strip()
glist = genre.split(',') # one movie can be in several genres
if 'Adventure' in glist: # check if movie belongs to genre Adventure
rating = float(cols[1].strip())
if rating > top[0]:
top = [rating,cols[6]]
fh.close()
print(top)
[8.9, 'The Lord of the Rings: The Return of the King']