Variant calling
From sequence data to variant call set
Per Unneberg
NBIS
15-Nov-2023
Yesterday
| 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9 | 10 | 11 | 12 | 13 | 14 | 15 |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| T | T | A | C | A | A | T | C | C | G | A | T | C | G | T |
| T | T | A | C | G | A | T | G | C | G | C | T | C | G | T |
| T | C | A | C | A | A | T | G | C | G | A | T | G | G | A |
| T | T | A | C | G | A | T | G | C | G | C | T | C | G | T |
| * | * | * | * | * | * |
\[ \begin{align} \pi & = \sum_{j=1}^S h_j = \sum_{j=1}^{S} \frac{n}{n-1}\left(1 - \sum_i p_i^2 \right) \\ & \stackrel{S=6,\\ n=4}{=} \sum_{j=1}^{6} \frac{4}{3}\left(1 - \sum_i p_i^2\right) \\ & = \frac{4}{3}\left(\mathbf{\color{#a7c947}{4}}\left(1-\frac{1}{16}- \frac{9}{16}\right) + \mathbf{\color{#a7c947}{2}}\left(1 - \frac{1}{4} - \frac{1}{4}\right)\right) = \frac{10}{3} \end{align} \]
\[ \begin{align} \pi & = \frac{\sum_{i=1}^{n-1}i(n-i)\xi_i}{n(n-1)/2} \\ & \stackrel{n=4}{=} \frac{1*(4-1)*4 + 2*(4-2)*2}{6} = \frac{10}{3} \end{align} \]
This is not how real data looks like from the beginning…
The real data
@SRR9309790.10003134
TAAATCGATTCGTTTTTGCTATCTTCGTCT
+
AAFFFJJJJJJJFJJJJJJJJJJJJJJJJJ
@SRR9309790.10003222
TAAATCGATTCGTTTTTGCTATCTTCGTCT
+
AAFFFJJJJJJJJJJJJJJJJJJJJJJJJJ30431 30441 30451 30461
CATTGGCAATGGCATCAGTTGAGCATCTTAGTACGAACTAAAAGCTG
...............M...............................
...............A...
...............A.................
..............................................
..............................................
..............................................
...............A...............................
...............A...............................
,,,,,,,,,,,,a,,a,,,,,,,,,,,,,,,,,,,,,,,,,,,,a,,
,,,,,,,,,,,,,,,a,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,
,,,,,,,,,,,,,,,a,,,,,,,,,,,,,,,,,,,,,,a,,,,,aa,
,,,,,,,,,,,,,,,a,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,
,,,,,,,,,,,,,a,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,
The process
Variant and genotype calling
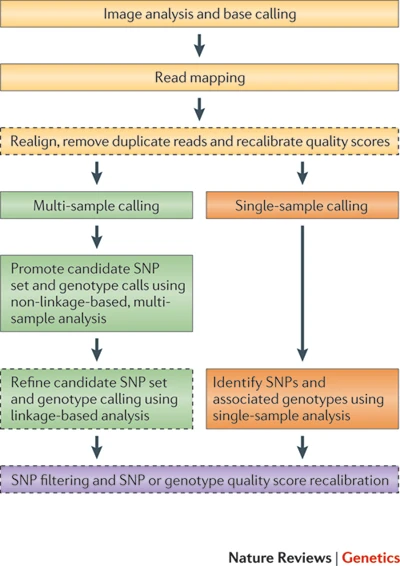
Nielsen et al. (2011)
- SNP calling
- Identification of polymorphic sites (\(>1\)% allele frequency)
- Variant calling
- Identification of variant sites (sufficient that any allele differs)
- Genotype calling
- Determine the allele combination for each individual (aa, aA, or AA for bi-allelic variants)
Knowing variant sites informs us of possible genotypes and improves genotyping.
Example: knowing a site has A or C limits possible genotype calls to AA, AC, or CC
Sequencing DNA
Sequencing technologies

Illumina NovaSeq 600
Scale up and down with a tunable output of up to 6 Tb and 20B single reads in < 2 days.
Up to 2X250 bp read length. Price example: 8,000 SEK total for resequencing 3Gbp genome to 30X
PacBio Revio
Up to 360 Gb of HiFi reads per day, equivalent to 1,300 human whole genomes per year.
Tens of kilobases long HiFi reads. Price example (Sequel II): ~35kSEK per library and SMRT cell
DNA sequences in FASTQ format
-rw-r--r-- 1 runner docker 302K Nov 15 11:10 PUN-Y-INJ_R1.fastq.gz
-rw-r--r-- 1 runner docker 393K Nov 15 11:10 PUN-Y-INJ_R2.fastq.gzFormat:
- sequence id (prefixed by
@) - DNA sequence
- separator (
+) - Phred base quality scores
DNA sequence quality control
Quality values represent the probability \(P\) that the call is incorrect. They are coded as Phred quality scores \(Q\). Here, \(Q=20\) implies 1% probability of error, \(Q=30\) 0.1% and so on. Typically you should not rely on quality values below \(20\).
\[ Q = -10 \log_{10} P \]
Sequencing approaches
Despite price drop, still need to make choices regarding depth and breadth of sequencing coverage and number of samples.
Genome assembly and population resequencing
Population resequencing

Read mapping
Sequence alignment maps reads to a reference
. characters is the consensus sequence. Bases are colored by nucleotide. Letter case indicates forward (upper-case) or reverse (lower-case) alignment. * is placeholder for deleted base.
Aim of sequence alignment (read mapping) is to determine source in reference sequence. Some commonly used read mappers for resequencing are
- BWA, BWA-MEM (H. Li, 2013)
- Novoalign (https://www.novocraft.com/)
- Minimap2 (H. Li, 2018)
For a recent comprehensive comparison see Donato et al. (2021)
Alignments are stored in BAM format
Header information
@HD VN:1.6 SO:coordinate
@SQ SN:LG4 LN:100000
@RG ID:SRR9309790 SM:PUN-Y-INJ PL:ILLUMINA
@PG ID:bwa PN:bwa VN:0.7.17-r1188 CL:bwa mem -R @RG\tID:SRR9309790\tSM:PUN-Y-INJ\tPL:ILLUMINA -p -t 14 -M tiny/M_aurantiacus_v1_splitline_ordered.fasta -Format: metadata record types prefixed with @, e.g., @RG is the read group
Alignments
SRR9309790.7750070 65 LG4 1 39 45S9M1I96M = 83782 83781 TGAACTATAGTCGATGGGACGAATACCCCCCTGAACTTGCGAAGGGGACAATTACCCCCCTCTGTTATGTTTCAGTCAATTTCATGTTTGATTTTTAGATTTTTAATTAATTATATATTTTTTGCAATTTGTAACCTCTTTAACCTTTATT AAFFFJJJJJJJJJJJJJJJJJJJJJJJJJJJJJJJJJJJJJJJJJJAJJJJJJJJJJJJJJJJJJJJJJJJJJJJJJFJJJJJJJJJJJFFJFJJJJJJJJJJFJJJJJJJJFJJJJJJJJJAJJJJJJJJJJJJJJJJJFA<FFJJ7FF SA:Z:LG4,83018,+,30M2I28M91S,37,5; MC:Z:12S10M2I123M4S MD:Z:16C28C59 PG:Z:MarkDuplicates RG:Z:SRR9309790 NM:i:3 MQ:i:50 AS:i:88 XS:i:70 ms:i:5185Some important columns: 1:QUERY, 3:REFERENCE, 4:POSITION, 5:MAPQ, 6:CIGAR. The CIGAR string compiles information on the alignment, such as match (M), soft clipping (S), and insertion to reference (I)
Mapped alignments can be viewed with samtools tview
30431 30441 30451 30461 30471
CATTGGCAATGGCATCAGTTGAGCATCTTAGTACGAACTAAAAGCTGCGAAAAAATATTT
...............M............................................
...............A... ,,,,,,,,,,
...............A................. ,,,,,,,,,,
.............................................. ,,,,,,,,
..............................................
..............................................
...............A............................................
...............A............................................
,,,,,,,,,,,,a,,a,,,,,,,,,,,,,,,,,,,,,,,,,,,,a,,a,,,,,,,,,,,,
,,,,,,,,,,,,,,,a,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,
,,,,,,,,,,,,,,,a,,,,,,,,,,,,,,,,,,,,,,a,,,,,aa,a,,,,,,,,,,,,
,,,,,,,,,,,,,,,a,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,
,,,,,,,,,,,,,a,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,
30431 30441 30451 30461 30471
CATTGGCAATGGCATCAGTTGAGCATCTTAGTACGAACTAAAAGCTGCGAAAAAATATTT
...............A............................................
,,,,,,, ..........................
...............A..... .....................
...............A...A.... .....................
...............A.........
...............A...........C.............
,,,,,,,,,,,,,,,a,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,
,,,,,,,,,,,,,,,a,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,
...............A............................................
,,,,,,,,,,,,,,,a,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,
,,,,,,,,,,,,,,,a,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,
,,,,,,,,,,,,,,,a,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,
,,,,,,,,,,,,,,,a,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,
...............A............................................
,,,g,,,,,,,a,,,a,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,
.............A............................................
Aka pileup format. Forward (.) and backward (,) mapping reads. Mismatches shown as letters.
Potential error corrections and pitfalls
Instrument
- PCR duplicates -> MarkDuplicates
- systematic errors from sequencing machine -> Base Quality Score Recalibration (BQSR)
Reference
- quality of reference sequence!
- repetitive sequence
Variant calling and genotyping
Variants show up in pileup alignments
Sample PUN-Y-INJ
30431 30441 30451 30461 30471
CATTGGCAATGGCATCAGTTGAGCATCTTAGTACGAACTAAAAGCTGCGAAAAAATATTT
...............M............................................
...............A... ,,,,,,,,,,
...............A................. ,,,,,,,,,,
.............................................. ,,,,,,,,
..............................................
..............................................
...............A............................................
...............A............................................
,,,,,,,,,,,,a,,a,,,,,,,,,,,,,,,,,,,,,,,,,,,,a,,a,,,,,,,,,,,,
,,,,,,,,,,,,,,,a,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,
,,,,,,,,,,,,,,,a,,,,,,,,,,,,,,,,,,,,,,a,,,,,aa,a,,,,,,,,,,,,
,,,,,,,,,,,,,,,a,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,
,,,,,,,,,,,,,a,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,
Sample PUN-R-ELF
30431 30441 30451 30461 30471
CATTGGCAATGGCATCAGTTGAGCATCTTAGTACGAACTAAAAGCTGCGAAAAAATATTT
...............A............................................
,,,,,,, ..........................
...............A..... .....................
...............A...A.... .....................
...............A.........
...............A...........C.............
,,,,,,,,,,,,,,,a,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,
,,,,,,,,,,,,,,,a,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,
...............A............................................
,,,,,,,,,,,,,,,a,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,
,,,,,,,,,,,,,,,a,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,
,,,,,,,,,,,,,,,a,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,
,,,,,,,,,,,,,,,a,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,
...............A............................................
,,,g,,,,,,,a,,,a,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,
.............A............................................
Potential variants show up as multiple mismatches in a column. Two questions arise:
- how do we detect variant sites?
- how do we distinguish variants from sequencing error?
Simple approach: filter bases on quality (e.g., Q20), call heterozygous if 20-80% bases non-reference.
Issues: undercalls heterozygotes, no measure of uncertainty
Solution: probabilistic methods!
We can calculate likelihoods of observed data
Example (exluding sequencing error!)
TGC
.K.
...
,,,
.T.
.T.
...
,,,
,t,
Goal: calculate likelihood of observing \(X=\)G,g,T,T,G,g,t from a genotype \(G\). We assume each observation \(X_i\) can be treated independently. We restrict possible genotypes to the observed alleles (i.e., G, T). Some observations:
Prob(\(X_1=\)G assuming genotype T,T) = P(\(X_1=\)G|T,T) = \(0\)
Prob(\(X_1=\)G assuming genotype G,G) = P(\(X_1=\)G|G,G) = \(1\)
Prob(\(X_1=\)G assuming genotype G,T) = P(\(X_1=\)G|G,T) = \(0.5\)
To get total likelihood \(P(X|G)\) assuming a genotype \(G\) (here G,T), we can multiply over all observations (reads):
\[ \begin{align} P(X|G,T) & = P(X_1=G|G,T)P(X_2=g|G,T)P(X_3=T|G,T)P(X_4=T|G,T) \\ & P(X_5=G|G,T)P(X_6=g|G,T)P(X_7=t|G,T) = 0.5^7 \end{align} \]
We can use Bayes’ theorem to genotype
Example
TGC
.K.
...
,,,
.T.
.T.
...
,,,
,t,
For a given site, we have a number of observations \(X\). We have shown we can calculate the likelihood of observing \(X\) given a genotype \(G\), \(P(X|G)\).
However; what we really want to know is the most likely genotype \(G\) given the data \(X\), or \(P(G|X)\).
Apply Bayes’ theorem:
\[ P(G|X) \sim P(X|G)\cdot P(G) \]
\[ \text{posterior} \sim \text{likelihood} \cdot \text{prior} \]
Consequently we need to set a prior on \(G\). If allele frequencies are known, we can constrain the frequencies; for example, if A is known to be low (\(\sim1\)%) AA genotype is very unlikely. Otherwise, could set all equal (flat prior).
Genotype likelihoods
We have outlined a probabilistic approach to variant calling where we obtain a posterior probability of observing a genotype \(G\) given data \(X\):
\[ P(G|X) \sim P(X|G)P(G) \]
Assuming a bi-allelic site, and letting \(H_1, H_2\) denote the two alleles, we have three possible genotype likelihoods \(P(H_1H_1|X)\), \(P(H_1H_2|X)\), and \(P(H_2H_2|X)\).
The highest posterior probability is typically chosen as the genotype call, with a measure confidence represented by the genotype probability or ratio between the two most probable calls.
Genotype likelihoods are often represented as Phred-scaled likelihoods (again!):
\[ \text{QUAL} = -10 \log_{10} P(G|X) \]
Variant Call Format (VCF) - header
bcftools view --header-only allsites.vcf.gz | head --lines 1
bcftools view --header-only allsites.vcf.gz | grep "##FILTER"
bcftools view -h allsites.vcf.gz | grep "##INFO" | head -n 4
bcftools view -h allsites.vcf.gz | grep "##FORMAT" | head -n 8##fileformat=VCFv4.2
##FILTER=<ID=PASS,Description="All filters passed">
##FILTER=<ID=LowQual,Description="Low quality">
##INFO=<ID=AC,Number=A,Type=Integer,Description="Allele count in genotypes, for each ALT allele, in the same order as listed">
##INFO=<ID=AF,Number=A,Type=Float,Description="Allele Frequency, for each ALT allele, in the same order as listed">
##INFO=<ID=AN,Number=1,Type=Integer,Description="Total number of alleles in called genotypes">
##INFO=<ID=BaseQRankSum,Number=1,Type=Float,Description="Z-score from Wilcoxon rank sum test of Alt Vs. Ref base qualities">
##FORMAT=<ID=AD,Number=R,Type=Integer,Description="Allelic depths for the ref and alt alleles in the order listed">
##FORMAT=<ID=DP,Number=1,Type=Integer,Description="Approximate read depth (reads with MQ=255 or with bad mates are filtered)">
##FORMAT=<ID=GQ,Number=1,Type=Integer,Description="Genotype Quality">
##FORMAT=<ID=GT,Number=1,Type=String,Description="Genotype">
##FORMAT=<ID=MIN_DP,Number=1,Type=Integer,Description="Minimum DP observed within the GVCF block">
##FORMAT=<ID=PGT,Number=1,Type=String,Description="Physical phasing haplotype information, describing how the alternate alleles are phased in relation to one another; will always be heterozygous and is not intended to describe called alleles">
##FORMAT=<ID=PID,Number=1,Type=String,Description="Physical phasing ID information, where each unique ID within a given sample (but not across samples) connects records within a phasing group">
##FORMAT=<ID=PL,Number=G,Type=Integer,Description="Normalized, Phred-scaled likelihoods for genotypes as defined in the VCF specification">FILTER defines applied filters , INFO fields provide additional information to genotypes, FORMAT specification fields define genotype entries, and more. NB: PL format definition.
Variant Call Format (VCF) - data
bcftools view --header-only --samples PUN-R-ELF,PUN-Y-INJ allsites.vcf.gz |\
tail --lines 1
bcftools view --no-header --samples PUN-R-ELF,PUN-Y-INJ allsites.vcf.gz LG4:6886#CHROM POS ID REF ALT QUAL FILTER INFO FORMAT PUN-R-ELF PUN-Y-INJ
LG4 6886 . C T 722.43 . AC=2;AF=0.222;AN=4;BaseQRankSum=0;DP=82;ExcessHet=1.8123;FS=1.309;InbreedingCoeff=-0.155;MLEAC=4;MLEAF=0.222;MQ=60;MQRankSum=0;QD=18.52;ReadPosRankSum=0.577;SOR=0.44 GT:AD:DP:GQ:PGT:PID:PL:PS 0/1:3,8:11:50:.:.:189,0,50:. 0/1:2,2:4:45:.:.:45,0,45:.QUAL: Phred-scaled quality score for Prob(ALT is wrong): \(722.43\) (\(p=10^{-Q/10}=5.7e-73\))
INFO field summarizes data for all samples. For instance:
- allele count 2 (
AC=2) - allele frequency minor allele 0.222 (
AF=0.222)
Variant Call Format (VCF) - data
#CHROM POS ID REF ALT QUAL FILTER INFO FORMAT PUN-R-ELF PUN-Y-INJ
LG4 6886 . C T 722.43 . AC=2;AF=0.222;AN=4;BaseQRankSum=0;DP=82;ExcessHet=1.8123;FS=1.309;InbreedingCoeff=-0.155;MLEAC=4;MLEAF=0.222;MQ=60;MQRankSum=0;QD=18.52;ReadPosRankSum=0.577;SOR=0.44 GT:AD:DP:GQ:PGT:PID:PL:PS 0/1:3,8:11:50:.:.:189,0,50:. 0/1:2,2:4:45:.:.:45,0,45:.
Genotypes (GT:AD:DP:GQ:PGT:PID:PL:PS)
PUN-R-ELF: 0/1:3,8:11:50:.:.:189,0,50:.
GT=0/1, AD=3,8 => 3 REF, 8 ALT, DP=11 => sequence depth = 11, PL=189,0,50
PUN-Y-INJ: 0/1:2,2:4:45:.:.:45,0,45:.
GT=0/1, AD=2,2 => 2 REF, 2 ALT, DP=4 => sequence depth = 4, PL=45,0,45
Relative genotype probabilities
Can convert Phred-scaled quality scores to probabilities as
\[ p = 10^{-Q/10} \]
For PUN-R-ELF the relative probabilities are \(10^{-189/10}\approx1.26e-9\), \(10^{0}=1\), \(10^{50}=10^{-5}\).
Interpretation: 0/1 10,000 times more likely than 1/1 (\(1/10^{-5}\))
TCT
.Y.
.T.
...
.T.
,,,
.T.
,,,
.T.
...
...
...
,t,
,t,
,t,
,t,
TCT
.Y.
.T.
...
,,,
,,,
,,,
.T.
:::::
::::::
GATK best practice
Pros
- Best practices
- Large documentation
- Variant quality score recalibration
Cons
- Human-centric - very slow runtime on genomes with many sequences
- Complicated setup
Alternative variant callers
ANGSD
For low-coverage sequencing. Doesn’t do explicit genotyping; most methods take genotype uncertainty into account.
Reference bias: plot no. hets vs coverage for real data, e.g., conifer
Exercise on variant calling
The monkeyflower system

From https://jgi.doe.gov/csp-2021-genomic-resources-for-mimulus/
Plants in the genus Mimulus inhabit highly variable habitats and are famous for their extraordinary ecological diversity. Mimulus is now a powerful system for ecological genomic studies, thanks to its experimental tractability, rapidly growing research community, and the JGI-generated reference genome for M. guttatus.
Learning outcomes
Follow GATK best practices to
- Perform qc on sequencing reads and interpret results
- Prepare reference for read mapping
- Map reads to reference
- Mark duplicates
- Perform raw variant calling to generate a set of sites to exclude from recalibration
- Perform base quality score recalibration
- Perform variant calling on base recalibrated data
- Do genotyping on all samples and combine results to a raw variant call set
Bibliography
Variant calling





