Simulation
Primer on the coalescent and forward simulation
Per Unneberg
NBIS
15-Nov-2023
Intended learning outcomes
Introduction to the coalescent and simulation in msprime, with two exercises on the coalescent and msprime. The students should be able to:
- Describe need for simulations
- Demonstrate the coalescent step by step
- Detail some properties of coalescent trees
- Define the site-frequency spectrum
- Describe the coalescent with recombination
- Perform simple simulations in msprime
The Wright-Fisher model and simulations
Recap
Model of a population describing genealogies under the following assumptions
- discrete and non-overlapping generations
- haploid individuals or two subpopulations (males and females)
- constant population size
- all individuals are equally fit
- population has no geographical or social structure
- no recombination
Forward simulation
The Wright-Fisher model and simulations
Mean reproductive success = 63.4%. Can show for large populations P(no descendants)=\(1 - e^{-1} \approx 0.632\)
Forward and backward simulation
Simulated nodes are filled with black. Genealogy of interest is highlighted in thick black lines.
Why do we want simulations anyway?
- Null model
- Generate neutral null distributions to compare with observed data
- Exploration
- Use to gain understanding and improve interpretation of mutational processes
- Mathematical complexity
- No analytical solutions for linked selection and the like \(\rightarrow\) must use simulations
- Benchmarking
- Use to generate test data with known properties on which to test and evaluate new methods
- Model training
- Generate training data for machine learning, e.g., Approximate Bayesian Computation (ABC) or Neural Networks (NNs)
The coalescent
Coalescent simulations
The coalescent simulates the genealogy of a sample of individuals on which mutations are “sprinkled” according to a Poisson process.
- Simulate ancestry (genealogy)
Coalescent simulations
The coalescent simulates the genealogy of a sample of individuals on which mutations are “sprinkled” according to a Poisson process.
- Simulate ancestry (genealogy)
- Simulate mutations
Coalescent simulations
The coalescent simulates the genealogy of a sample of individuals on which mutations are “sprinkled” according to a Poisson process.
- Simulate ancestry (genealogy)
- Simulate mutations
Exercise
How many mutations are common to all samples? How many mutations does sample 1 have? Sample 2?
Assuming the ancestral state is denoted 0 (prior to the first generation) and the derived state 1, what are the sequences of the samples?
Simulating genealogies (Hahn, 2019, p. 115)
- Start with \(i=n\) chromosomes
Simulating genealogies (Hahn, 2019, p. 115)
Start with \(i=n\) chromosomes
Choose time to next coalescent event from an exponential distribution with parameter \(\lambda=i(i-1)/2\)
Simulating genealogies (Hahn, 2019, p. 115)
Start with \(i=n\) chromosomes
Choose time to next coalescent event from an exponential distribution with parameter \(\lambda=i(i-1)/2\)
Choose two chromosomes at random to coalesce
Simulating genealogies (Hahn, 2019, p. 115)
Start with \(i=n\) chromosomes
Choose time to next coalescent event from an exponential distribution with parameter \(\lambda=i(i-1)/2\)
Choose two chromosomes at random to coalesce
Merge the two lineages and set \(i \rightarrow i - 1\)
Simulating genealogies (Hahn, 2019, p. 115)
- Start with \(i=n\) chromosomes
- Choose time to next coalescent event from an exponential distribution with parameter \(\lambda=i(i-1)/2\)
- Choose two chromosomes at random to coalesce
- Merge the two lineages and set \(i \rightarrow i - 1\)
- If \(i>1\), go to step 2; if not, stop.
Simulating genealogies (Hahn, 2019, p. 115)
- Start with \(i=n\) chromosomes
- Choose time to next coalescent event from an exponential distribution with parameter \(\lambda=i(i-1)/2\)
- Choose two chromosomes at random to coalesce
- Merge the two lineages and set \(i \rightarrow i - 1\)
- If \(i>1\), go to step 2; if not, stop.
Simulating genealogies (Hahn, 2019, p. 115)
- Start with \(i=n\) chromosomes
- Choose time to next coalescent event from an exponential distribution with parameter \(\lambda=i(i-1)/2\)
- Choose two chromosomes at random to coalesce
- Merge the two lineages and set \(i \rightarrow i - 1\)
- If \(i>1\), go to step 2; if not, stop.
Simulating genealogies (Hahn, 2019, p. 115)
- Start with \(i=n\) chromosomes
- Choose time to next coalescent event from an exponential distribution with parameter \(\lambda=i(i-1)/2\)
- Choose two chromosomes at random to coalesce
- Merge the two lineages and set \(i \rightarrow i - 1\)
- If \(i>1\), go to step 2; if not, stop.
Simulating genealogies (Hahn, 2019, p. 115)
- Start with \(i=n\) chromosomes
- Choose time to next coalescent event from an exponential distribution with parameter \(\lambda=i(i-1)/2\)
- Choose two chromosomes at random to coalesce
- Merge the two lineages and set \(i \rightarrow i - 1\)
- If \(i>1\), go to step 2; if not, stop.
Simulating genealogies (Hahn, 2019, p. 115)
- Start with \(i=n\) chromosomes
- Choose time to next coalescent event from an exponential distribution with parameter \(\lambda=i(i-1)/2\)
- Choose two chromosomes at random to coalesce
- Merge the two lineages and set \(i \rightarrow i - 1\)
- If \(i>1\), go to step 2; if not, stop.
Simulating genealogies (Hahn, 2019, p. 115)
- Start with \(i=n\) chromosomes
- Choose time to next coalescent event from an exponential distribution with parameter \(\lambda=i(i-1)/2\)\(^1\)
- Choose two chromosomes at random to coalesce
- Merge the two lineages and set \(i \rightarrow i - 1\)
- If \(i>1\), go to step 2; if not, stop.
\(^1\): The exponential can be parametrized in two different ways, so that the parameter to the function is either \(\lambda\) or \(\beta=1/\lambda\).
Simulating genealogies (Hahn, 2019, p. 115)
- Start with \(i=n\) chromosomes
- Choose time to next coalescent event from an exponential distribution with parameter \(\lambda=i(i-1)/2\)\(^1\)
- Choose two chromosomes at random to coalesce
- Merge the two lineages and set \(i \rightarrow i - 1\)
- If \(i>1\), go to step 2; if not, stop.
\(^1\): The exponential can be parametrized in two different ways, so that the parameter to the function is either \(\lambda\) or \(\beta=1/\lambda\).
Some properties of the tree
Expected waiting time to coalesce when \(i\) lineages: \(E(T_i) = \frac{2}{i(i-1)}\)
Branch lengths can be derived from waiting times. For instance, \(\tau_1=\tau_2=T_5+T_4\) and \(\tau_4=\tau_5=T_5\)
Time to the most recent common ancestor (MRCA) is sum of wating times: \(T_{MRCA} = \sum_{i=2}^n T_i\)
with expected value \(E(T_{MRCA}) = \sum_{i=2}^nE(T_i) = 2\left(1 - \frac{1}{n}\right)\)
The expected total tree height is \(E(T_{total}) = \sum_{i=2}^n iE(T_i) = 2\sum_{i=2}^n\frac{1}{i-1}\)
Coalescent simulations vary in topology and height
Examples of coalescent simulations. Note the variation in tree topology and height.
Diminishing returns of adding more samples
Adding mutations
Mutations are added by placing them on branches, where the probability of ending up on a branch \(\tau_i\) is equal to its normalized length, where normalization is by the total tree branch length:
\[ P(\text{mutation on branch }i) = \frac{\tau_i}{\sum_j\tau_j} = \frac{\tau_i}{T_{total}} \]
The total number of segregating sites \(S\) to be thrown on the tree is modelled as a Poisson random variable which expresses the probability of a given number of events in time \(t\):
\[ S = Po(\theta/2T_{total}) \]
The coalescent and diversity
| 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9 | 10 | 11 | 12 | 13 | 14 | 15 |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| T | T | A | C | A | A | T | C | C | G | A | T | C | G | T |
| T | T | A | C | G | A | T | G | C | G | C | T | C | G | T |
| T | C | A | C | A | A | T | G | C | G | A | T | G | G | A |
| T | T | A | C | G | A | T | G | C | G | C | T | C | G | T |
| * | * | * | * | * | * |
\[ \begin{align} \pi & = \sum_{j=1}^S h_j = \sum_{j=1}^{S} \frac{n}{n-1}\left(1 - \sum_i p_i^2 \right) \\ & \stackrel{S=6,\\ n=4}{=} \sum_{j=1}^{6} \frac{4}{3}\left(1 - \sum_i p_i^2\right) \\ & = \frac{4}{3}\left(\mathbf{\color{#a7c947}{4}}\left(1-\frac{1}{16}-\frac{9}{16}\right) + \mathbf{\color{#a7c947}{2}}\left(1 - \frac{1}{4} - \frac{1}{4}\right)\right) = \frac{10}{3} \end{align} \]
In this notation one can show that \(\pi\), the nucleotide diversity, is
\[ \begin{align} \pi & = \frac{\sum_{i=1}^{n-1}i(n-i)\xi_i}{n(n-1)/2} \\ & \stackrel{n=4}{=} \frac{1*(4-1)*4 + 2*(4-2)*2}{6} = \frac{10}{3} \end{align} \]
Many statistical quantities can be related to the site frequency spectrum (SFS), which is a summary of the frequencies of the segregating sites. Let \(\xi_i\) be the number of chromosomes in the sample with \(i\) minor alleles. In the example above we have \(S=6\) mutations on \(n=4\) chromosomes.
The impact of topology on the SFS
Exercise on the coalescent
The coalescent with recombination
On non-recombining chromosomes and assortment
Both siblings inherit chromosome from paternal grandfather
Chromosomes coalesce at father
Siblings inherit different grandparental chromosomes \(\Rightarrow\) chromosomes coalesce God knows when in the past
Genealogies differ
The ancestral recombination graph



Y. C. Brandt et al. (2022, fig. 1a)
Properties:
- marginal trees constitute a sequence of trees (tree sequence) along a chromosome
- each tree represents the genealogy of a non-recombining part of the chromosome
- neighbouring trees are correlated
Interpretation: chromosomes are mosaics of non-recombining units
The ancestral recombination graph as sequence of trees

Properties:
- marginal trees constitute a sequence of trees (tree sequence) along a chromosome
- each tree represents the genealogy of a non-recombining part of the chromosome
- neighbouring trees are correlated
Interpretation: chromosomes are mosaics of non-recombining units
msprime is a fast coalescent simulator
Original coalescent simulator implementions assumed small sample sizes and could only simulate short recombining sequences. msprime was developed to address these shortcomings as today we have
- chromosome-level assemblies that require full treatment of recombination (very complex problem)
- biobank datasets consisting of 100’s of thousands of samples
Features
- can simulate millions of whole chromosomes
- well-designed mature API
- succinct data structure (tree sequences) has led to advances in forwards-time simulation
- can be used together with forwards-time simulators
Limitations
- assumes neutrality - can’t simulate selection
msprime stores variation data as tree sequences

Tree sequences (Baumdicker et al., 2022, fig. 2)
Tree sequences compress data

Data compression (Kelleher et al., 2019, fig. 1c)
Simulating ancestry with msprime
From msprime quickstart
import msprime
# Simulate an ancestral history for 3 diploid samples under the coalescent
# with recombination on a 5kb region with human-like parameters.
ts = msprime.sim_ancestry(
samples=3,
recombination_rate=1e-8,
sequence_length=5_000,
population_size=10_000,
random_seed=123456)
print(ts.draw_svg())Simulating mutations with msprime
msprime exercise
Forward simulation in SLiM
SLiM
SLiM (Selection on Linked Mutations) (Haller & Messer, 2019) is a forwards-time simulator. As its name implies, it models selection and thus is a complement to coalescent-based simulators.
Why SLiM?
- flexibility - scripting language Eidos allows for modelling complex scenarios with little code
- performance - optimized code base
- GUI - interactive execution and graphical debugging
Haller & Messer (2022)

Forward simulation in SLiM
- Execution of
first()events - Execution of
early()events - Generation of offspring; for each offspring:
- Choose source subpop for parental individuals, based on migration rates
- Choose parent 1, based on cached fitness values
- Choose parent 2, based on fitness and any defined
mateChoice()callbacks - Generate the candidate offspring, with mutation and recombination (including
mutation()andrecombination()callbacks) - Suppress/modify the candidate, using defined
modifyChild()callbacks
- Removal of fixed mutations unless
convertToSubstitution==F - Offspring become parents
- Execution of
late()events - Fitness recalculation using
mutationEffect()andfitnessEffect()callbacks - Tick/cycle count increment
Simple neutral simulation in SLiM
// set up a simple neutral simulation
initialize()
{
// set the overall mutation rate
initializeMutationRate(1e-7);
// m1 mutation type: neutral
initializeMutationType("m1", 0.5, "f", 0.0);
// g1 genomic element type: uses m1 for all mutations
initializeGenomicElementType("g1", m1, 1.0);
// uniform chromosome of length 100 kb
initializeGenomicElement(g1, 0, 99999);
// uniform recombination along the chromosome
initializeRecombinationRate(1e-8);
}
// create a population of 500 individuals
1 early()
{
sim.addSubpop("p1", 500);
}
// run to tick 10000
10000 early()
{
sim.simulationFinished();
}Comes with a huge manual (Haller & Messer, 2022) (700+ pages!)
Recapitation - combining the best of two worlds
A recent addition to SLiM is that it records data in tree sequence format (as in msprime) we can combine backward and forward simulations!
Backward simulation that adds coalescent (neutral) history
Forward simulation with selection or some other process that isn’t supported by the coalescent
Applications
Simulation of null distributions

Authors inferred demographic history and recombination map. The inferred values were used to simulate data with msprime. SweepFinder2 (DeGiorgio et al., 2016) was used to calculate composite likelihood ratio (CLR) scores on both observed and simulated data to assess significance quantiles.
Simulations of genomic landscapes in monkeyflower
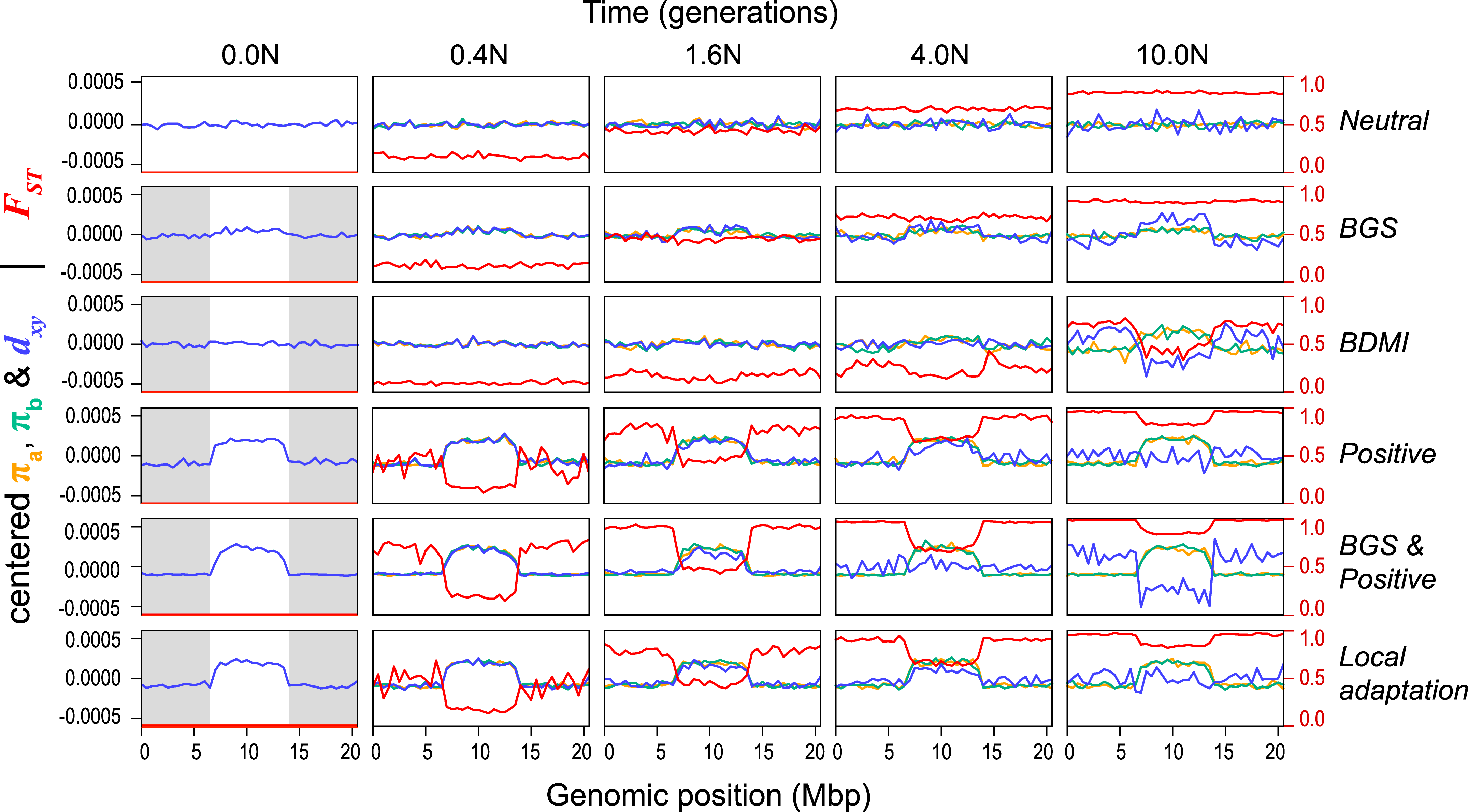
Monkeyflower (Mimulus aurantiacus) radiation in California shows wide adaptive range and phenotypic diversity. Stankowski et al. (2019) et al use simulations (SLiM and msprime) to shed light on processes that shape genomic landscapes, which are correlated between species.
We will look more closely at this system tomorrow.
Neural network recombination landscape prediction

Use msprime simulations to generate training data for neural network.
Bibliography
Simulation primer
