DNA sequencing data
DNA sequencing approaches and data
Sequencing technologies
Illumina NovaSeq 600
Scale up and down with a tunable output of up to 6 Tb and 20B single reads in < 2 days.
Up to 2X250 bp read length. Price example: 8,000 SEK total for resequencing 3Gbp genome to 30X
PacBio Revio
Up to 360 Gb of HiFi reads per day, equivalent to 1,300 human whole genomes per year.
Tens of kilobases long HiFi reads. Price example (Sequel II): ~35kSEK per library and SMRT cell
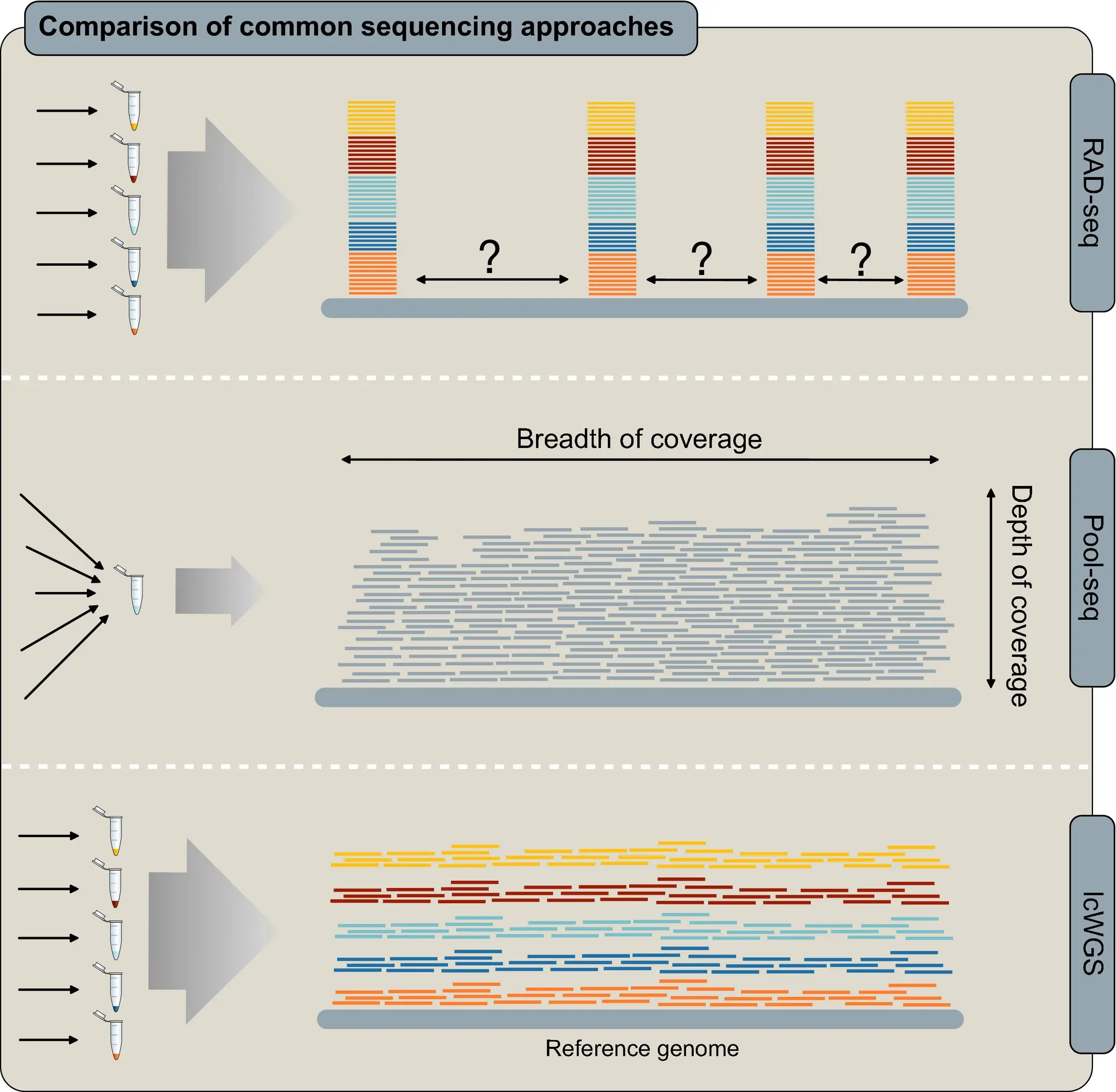
Sequencing approaches
Despite price drop, still need to make choices regarding depth and breadth of sequencing coverage and number of samples.
Lou et al. (2021)
Our focus will be on Whole Genome reSequencing (WGS), mostly high-coverage.
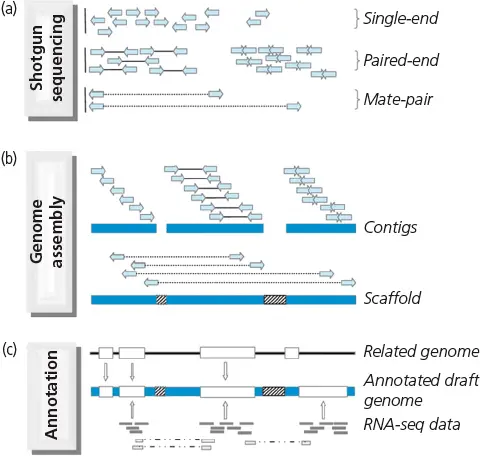
Genome assembly and population resequencing
Genome assembly
Allendorf et al. (2022)
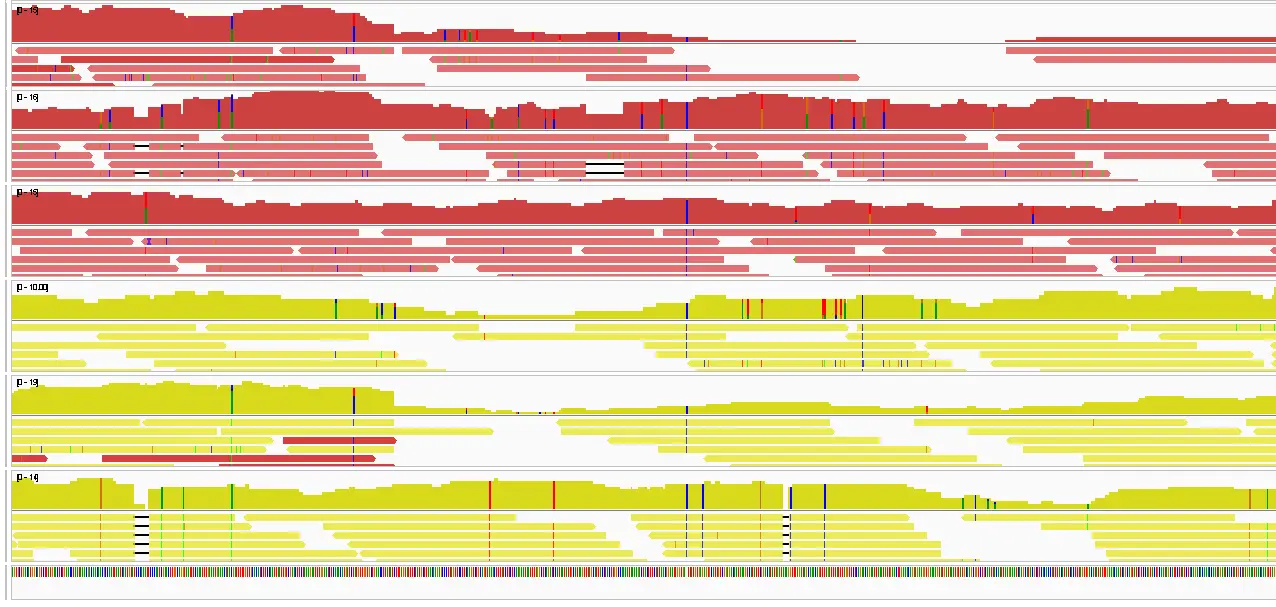
Population resequencing
DNA sequences in FASTQ format
-rw-r--r-- 1 runner runner 302K Sep 18 22:10 fastq/PUN-Y-INJ_R1.fastq.gz
-rw-r--r-- 1 runner runner 393K Sep 18 22:10 fastq/PUN-Y-INJ_R2.fastq.gzFormat:
- sequence id (prefixed by
@) - DNA sequence
- separator (
+) - Phred base quality scores
DNA sequence quality control
Quality values represent the probability P that the call is incorrect. They are coded as Phred quality scores Q. Here, Q=20 implies 1% probability of error, Q=30 implies 0.1% and so on. Typically you should not rely on quality values below 20.
Q = -10 \log_{10} P
A common way to do QC is with fastqc:
- For Illumina paired-end sequencing, the second read pair usually shows a larger drop in quality towards ends
- Trimming the sequences for adapter sequence and quality is good practice (e.g., with CutAdapt (Martin, 2011))
Bibliography
DNA data