// set up a simple neutral simulation
initialize()
{
// set the overall mutation rate
initializeMutationRate(1e-7);
// m1 mutation type: neutral
initializeMutationType("m1", 0.5, "f", 0.0);
// g1 genomic element type: uses m1 for all mutations
initializeGenomicElementType("g1", m1, 1.0);
// uniform chromosome of length 100 kb
initializeGenomicElement(g1, 0, 99999);
// uniform recombination along the chromosome
initializeRecombinationRate(1e-8);
}
// create a population of 500 individuals
1 early()
{
sim.addSubpop("p1", 500);
}
// run to tick 10000
10000 early()
{
sim.simulationFinished();
}Brief introduction to simulation packages and stdpopsim
Making simulations with stdpopsim, the standard library for population genetic simulation models
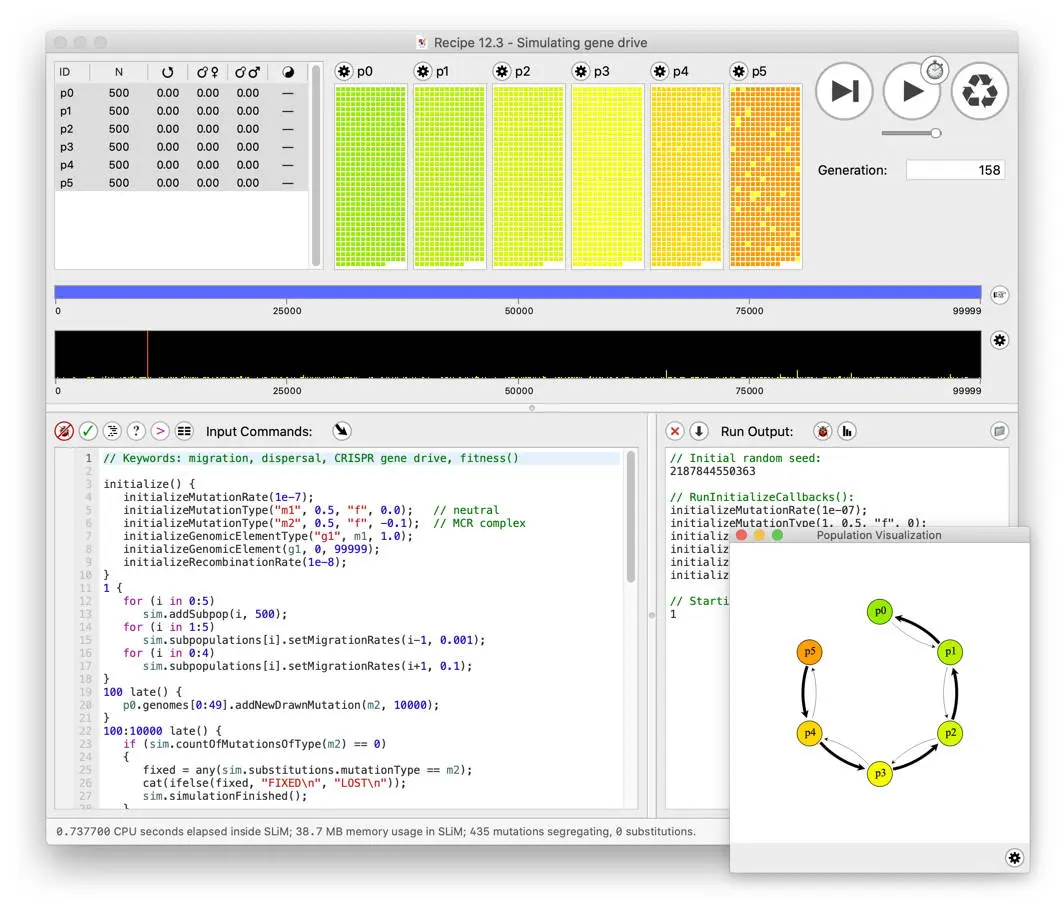
SLiM
Eidos: an R-like programming language
Comes with a huge manual (Haller & Messer, 2022) (700+ pages!)
SLiM resources
An introduction to SLiM (Ben Haller): video (1 hour)
13 March 2024: This is a one-hour introductory talk about SLiM, first presented during the Modeling & Theory in Population Biology series in 2024. It’s a great place to start if you’re not sure what SLiM is, what it can do, and why you might want to use it.
A practical Introduction to the Tree Sequence (Peter Ralph): video (24 min.)
16 June 2023: This is a fairly short talk about the tree sequence data structure, both in the abstract and as used in SLiM simulations. It’ll help you understand what tree sequences are, why you might want to use them, and how to get started with them.
https://messerlab.org/slim/#Videos
SLiM Workshops
We run 5-day SLiM workshops that are free and open to the public (with registration). The best place to watch for announcements of new workshops is the slim-discuss mailing list. We have one scheduled in Europe for 2026 so far, and expect that there will be several more:
17 August – 21 August 2026, University of Iceland, Reykjavík, Iceland
If you would like to host a workshop at your institution, please contact us!
The workshop materials are now online, for people who want to do it themselves; visit the online workshop download page for more information.


msprime
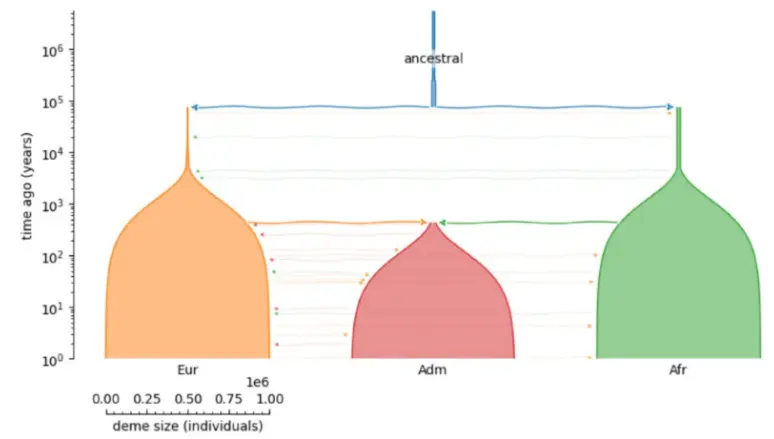
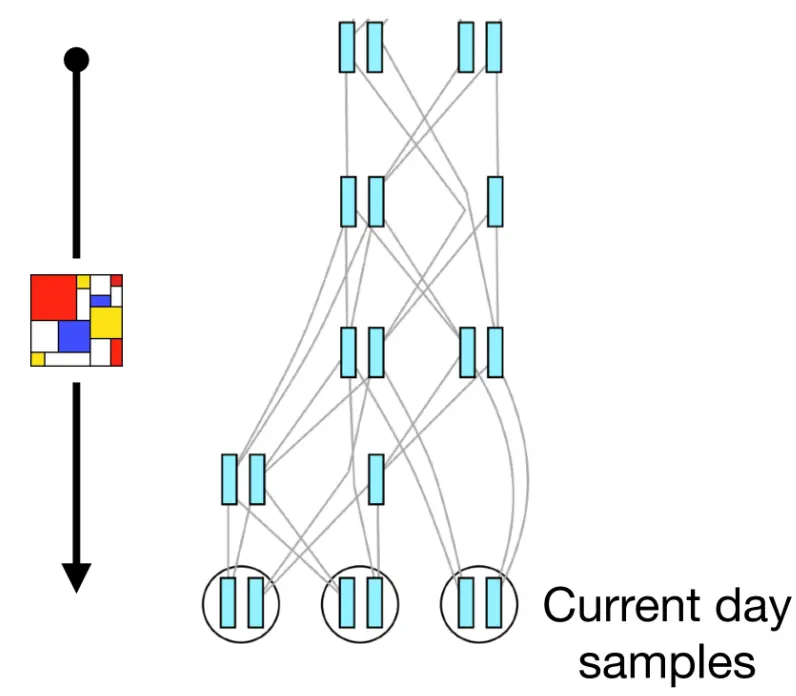
Recapitation

Recapitation
- Combine forward- & backward-time simulations
- E.g., only forward-simulate the period of interest (reduce burn-in)
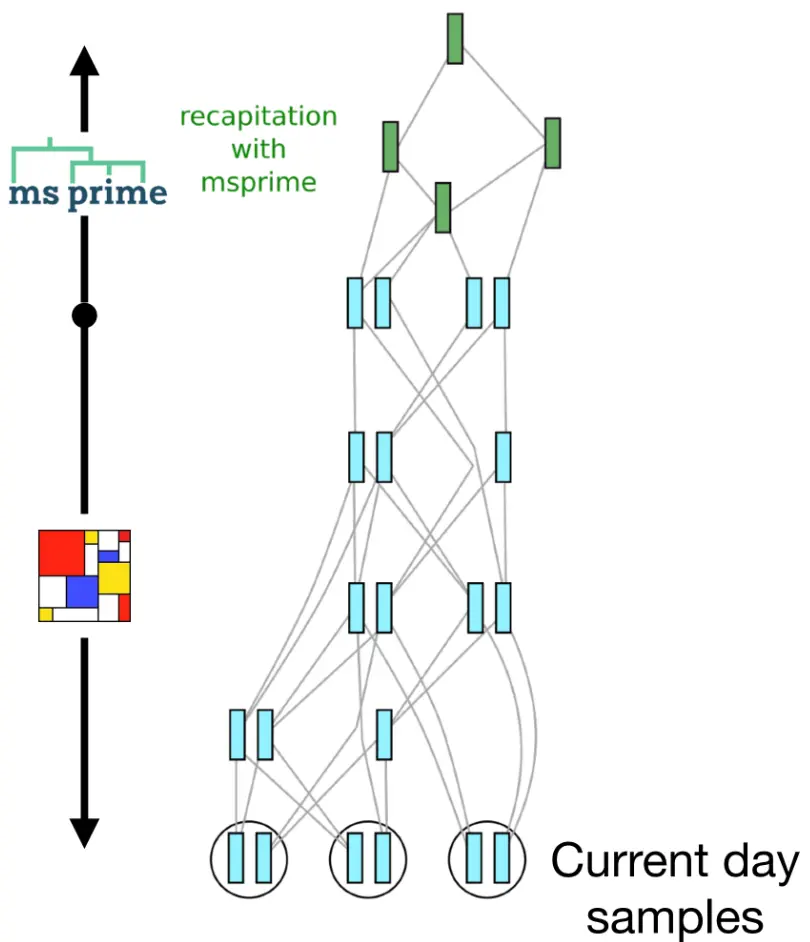
Recapitation

Recapitation
- Combine forward- & backward-time simulations
- E.g., only forward-simulate the period of interest (reduce burn-in)
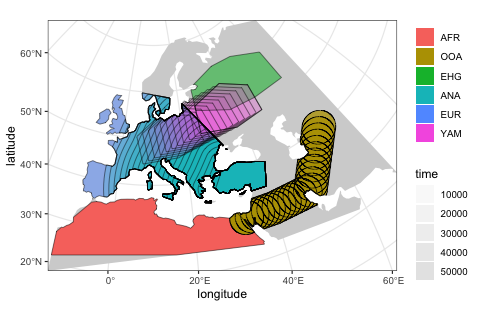
Spatial population genetics in R
slendr: Population Genetic Simulations in R
Overview
slendr is a toolbox for running population genomic simulations entirely in R. Our original motivation for developing it was to provide a framework for simulating spatially-explicit genomic data on real geographic landscapes, however, it has grown to be much more than that since then: slendr can now simulate data from traditional, non-spatial demographic models using msprime as a simulation engine, and it even supports selection scenarios via user-defined SLiM estension snippets.

PopSim consortium
stdpopsim
stdpopsim - specifying the engine
Bibliography
Simulation packages