Primer on the coalescent and forward simulation
Intended learning outcomes
Introduction to the coalescent and simulation in msprime, with two exercises on the coalescent and msprime. The students should be able to:
- Describe need for simulations
- Demonstrate the coalescent step by step
- Detail some properties of coalescent trees
- Define the site-frequency spectrum
- Describe the coalescent with recombination
- Perform simple simulations in msprime
The Wright-Fisher model and simulations
Recap
Model of a population describing genealogies under the following assumptions
- discrete and non-overlapping generations
- haploid individuals or two subpopulations (males and females)
- constant population size
- all individuals are equally fit
- population has no geographical or social structure
- no recombination
Forward simulation
The Wright-Fisher model and simulations
Forward and backward simulation
Simulated nodes are filled with black. Genealogy of interest is highlighted in thick black lines.
Why do we want simulations anyway?
- Null model
- Generate neutral null distributions to compare with observed data
- Exploration
- Use to gain understanding and improve interpretation of mutational processes
- Mathematical complexity
- No analytical solutions for linked selection and the like \rightarrow must use simulations
- Benchmarking
- Use to generate test data with known properties on which to test and evaluate new methods
- Model training
- Generate training data for machine learning, e.g., Approximate Bayesian Computation (ABC) or Neural Networks (NNs)
The coalescent
Coalescent simulations
Coalescent simulations
Coalescent simulations
The coalescent simulates the genealogy of a sample of individuals on which mutations are “sprinkled” according to a Poisson process.
- Simulate ancestry (genealogy)
- Simulate mutations
Exercise
How many mutations are common to all samples? How many mutations does sample 1 have? Sample 2?
Assuming the ancestral state is denoted 0 (prior to the first generation) and the derived state 1, what are the sequences of the samples?
Simulating genealogies (Hahn, 2019, p. 115)
Simulating genealogies (Hahn, 2019, p. 115)
Simulating genealogies (Hahn, 2019, p. 115)
Simulating genealogies (Hahn, 2019, p. 115)
Simulating genealogies (Hahn, 2019, p. 115)
Simulating genealogies (Hahn, 2019, p. 115)
Simulating genealogies (Hahn, 2019, p. 115)
Simulating genealogies (Hahn, 2019, p. 115)
Simulating genealogies (Hahn, 2019, p. 115)
Simulating genealogies (Hahn, 2019, p. 115)
Simulating genealogies (Hahn, 2019, p. 115)
- Start with i=n chromosomes
- Choose time to next coalescent event from an exponential distribution with parameter \lambda=i(i-1)/2^1
- Choose two chromosomes at random to coalesce
- Merge the two lineages and set i \rightarrow i - 1
- If i>1, go to step 2; if not, stop.
^1: The exponential can be parametrized in two different ways, so that the parameter to the function is either \lambda or \beta=1/\lambda.
Simulating genealogies (Hahn, 2019, p. 115)
- Start with i=n chromosomes
- Choose time to next coalescent event from an exponential distribution with parameter \lambda=i(i-1)/2^1
- Choose two chromosomes at random to coalesce
- Merge the two lineages and set i \rightarrow i - 1
- If i>1, go to step 2; if not, stop.
^1: The exponential can be parametrized in two different ways, so that the parameter to the function is either \lambda or \beta=1/\lambda.
Some properties of the tree
The waiting time for coalescence is shorter when there are more lineages
Time to the most recent common ancestor (MRCA) (i.e., the tree height) is the sum of the waiting times (T_2 + T_3 + ...)
On average, the time for coalescence when there are two remaining lineages is half the tree height
Coalescent simulations vary in topology and height
Examples of coalescent simulations. Note the variation in tree topology and height.
Diminishing returns of adding more samples
Adding mutations
Mutations are added by “throwing” them on branches, where the probability of ending up on a given branch is equal to its relative length.
The total number of segregating sites S to be thrown on the tree is modelled as a Poisson random variable which expresses the probability of a given number of events in a given time. Here the time is the total branch length of the tree and mutations are added with intensity proportional to the population mutation rate \theta.
The coalescent and diversity
| 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9 | 10 | 11 | 12 | 13 | 14 | 15 |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| T | T | A | C | A | A | T | C | C | G | A | T | C | G | T |
| T | T | A | C | G | A | T | G | C | G | C | T | C | G | T |
| T | C | A | C | A | A | T | G | C | G | A | T | G | G | A |
| T | T | A | C | G | A | T | G | C | G | C | T | C | G | T |
| * | * | * | * | * | * |
\begin{align} \pi & = \sum_{j=1}^S h_j = \sum_{j=1}^{S} \frac{n}{n-1}\left(1 - \sum_i p_i^2 \right) \\ & \stackrel{S=6,\\ n=4}{=} \sum_{j=1}^{6} \frac{4}{3}\left(1 - \sum_i p_i^2\right) \\ & = \frac{4}{3}\left(\mathbf{\color{#a7c947}{4}}\left(1-\frac{1}{16}-\frac{9}{16}\right) + \mathbf{\color{#a7c947}{2}}\left(1 - \frac{1}{4} - \frac{1}{4}\right)\right) = \frac{10}{3} \end{align}
Many statistical quantities can be related to the site frequency spectrum (SFS), which is a summary of the frequencies of the segregating sites. Let \xi_i be the number of chromosomes in the sample with i minor alleles. In the example above we have S=6 mutations on n=4 chromosomes.
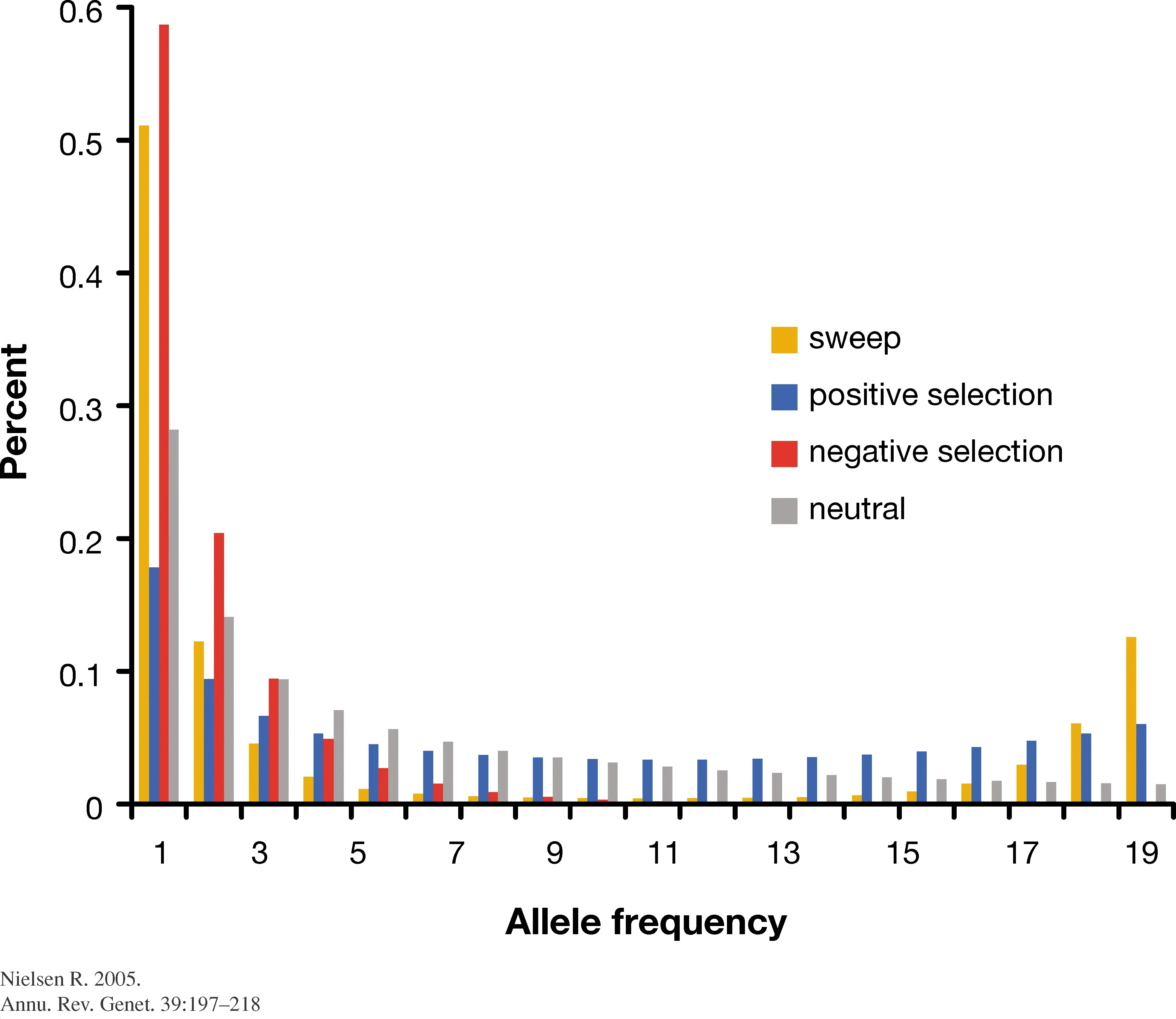
The impact of topology on the SFS
msprime
msprime stores data as succinct tree sequences
Tree sequences compress data and speedup analyses
- Compact storage (“domain specific compression”)
- Fast, efficient analysis (a “succinct” structure)
- Well tested, open source (active dev community)
Data compression
- Built-in functionality (well documented: http://tskit.dev)
…but limited support for major genomic rearrangements (e.g. inversions, large indels): genomes should be (reasonably) aligned => current primary focus = population genetics
Speed
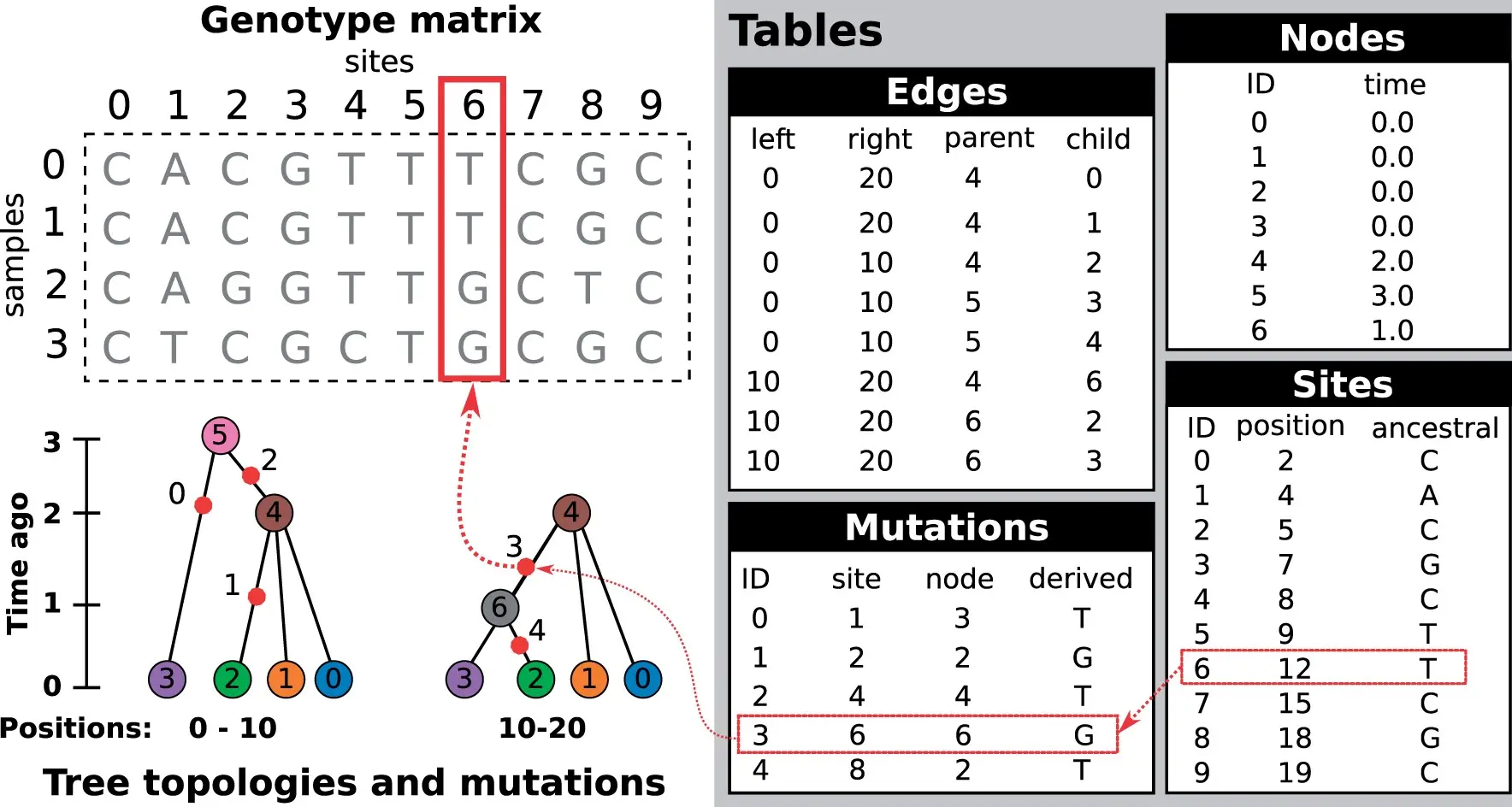
tskit terminology: the basics
- Multiple local trees exist along a genome of fixed length (by convention measured in base pairs)
- Genomes exist at specific times, and arerepresented by nodes (the same node can persist across many local trees)
- Some nodes are most recent common ancestors (MRCAs) of other nodes
- Entities are zero-based: the rst node has id 0, the second id 1, …
Images from online tutorial “Terminology & concepts” https://tskit.dev/tutorials/terminology_and_concepts.html
tskit terminology: nodes and edges
Nodes (=genomes)
- exist at a specific time
- can be flagged as “samples”
- can belong to “individuals” (e.g., 2 nodes per individuals in humans) and, if useful, “populations”
| id | flags | population | individual | time | metadata |
|---|---|---|---|---|---|
| 0 | 1 | 0 | 0 | 0.00000000 | |
| 1 | 1 | 0 | 0 | 0.00000000 | |
| 2 | 1 | 0 | 1 | 0.00000000 | |
| 3 | 1 | 0 | 1 | 0.00000000 | |
| 4 | 1 | 0 | 2 | 0.00000000 | |
| 5 | 1 | 0 | 2 | 0.00000000 | |
| 6 | 0 | 0 | -1 | 14.70054184 | |
| 7 | 0 | 0 | -1 | 40.95936939 | |
| 8 | 0 | 0 | -1 | 72.52965866 | |
| 9 | 0 | 0 | -1 | 297.22307150 | |
| 10 | 0 | 0 | -1 | 340.15496436 | |
| 11 | 0 | 0 | -1 | 605.35907657 |
Edges
- Connect a parent & child
- Have a left & right genomic coordinate
- Usually span multiple trees (e.g., edges connecting nodes 1+7 and 4+7)
| id | left | right | parent | child | metadata |
|---|---|---|---|---|---|
| 0 | 0 | 1000 | 6 | 2 | |
| 1 | 0 | 1000 | 6 | 5 | |
| 2 | 0 | 1000 | 7 | 1 | |
| 3 | 0 | 1000 | 7 | 4 | |
| 4 | 0 | 1000 | 8 | 3 | |
| 5 | 0 | 1000 | 8 | 6 | |
| 6 | 307 | 1000 | 9 | 0 | |
| 7 | 307 | 1000 | 9 | 7 | |
| 8 | 0 | 307 | 10 | 0 | |
| 9 | 0 | 567 | 10 | 8 | |
| 10 | 307 | 567 | 10 | 9 | |
| 11 | 0 | 307 | 11 | 7 | |
| 12 | 567 | 1000 | 11 | 8 | |
| 13 | 567 | 1000 | 11 | 9 | |
| 14 | 0 | 307 | 11 | 10 |
tskit terminology: sites and mutations
This is how we can encode genetic variation. Most genomic positions do not vary between genomes: usually we don’t bother tracking these.
tskit terminology: sites and mutations
We can create a site at a given genomic position with a fixed ancestral state.
| id | position | ancestral_state | metadata |
|---|---|---|---|
| 0 | 52 | C | |
| 1 | 200 | A | |
| 2 | 335 | A | |
| 3 | 354 | A | |
| 4 | 474 | G | |
| 5 | 523 | A | |
| 6 | 774 | C | |
| 7 | 796 | C | |
| 8 | 957 | A |
This is how we can encode genetic variation. Most genomic positions do not vary between genomes: usually we don’t bother tracking these.
tskit terminology: sites and mutations
We can create a site at a given genomic position with a fixed ancestral state.
| id | position | ancestral_state | metadata |
|---|---|---|---|
| 0 | 52 | C | |
| 1 | 200 | A | |
| 2 | 335 | A | |
| 3 | 354 | A | |
| 4 | 474 | G | |
| 5 | 523 | A | |
| 6 | 774 | C | |
| 7 | 796 | C | |
| 8 | 957 | A |
This is how we can encode genetic variation. Most genomic positions do not vary between genomes: usually we don’t bother tracking these.
Normally, a site is created in order to place one or more mutations at that site
| id | site | node | time | derived_state | parent | metadata |
|---|---|---|---|---|---|---|
| 0 | 0 | 8 | 247.85988972 | T | -1 | |
| 1 | 1 | 0 | 169.80687857 | C | -1 | |
| 2 | 2 | 3 | 31.84262397 | C | -1 | |
| 3 | 3 | 9 | 326.26095349 | C | -1 | |
| 4 | 3 | 7 | 71.04212649 | T | 3 | |
| 5 | 4 | 3 | 42.72352948 | C | -1 | |
| 6 | 5 | 7 | 55.44045835 | T | -1 | |
| 7 | 6 | 0 | 259.82567754 | T | -1 | |
| 8 | 7 | 8 | 169.87040769 | G | -1 | |
| 9 | 8 | 0 | 42.47396523 | C | -1 |
Bibliography
Population Genomics in Practice