Ancestral recombination graph inference
Inferring tree sequences for population genomics
What are Ancestral Recombination Graphs?
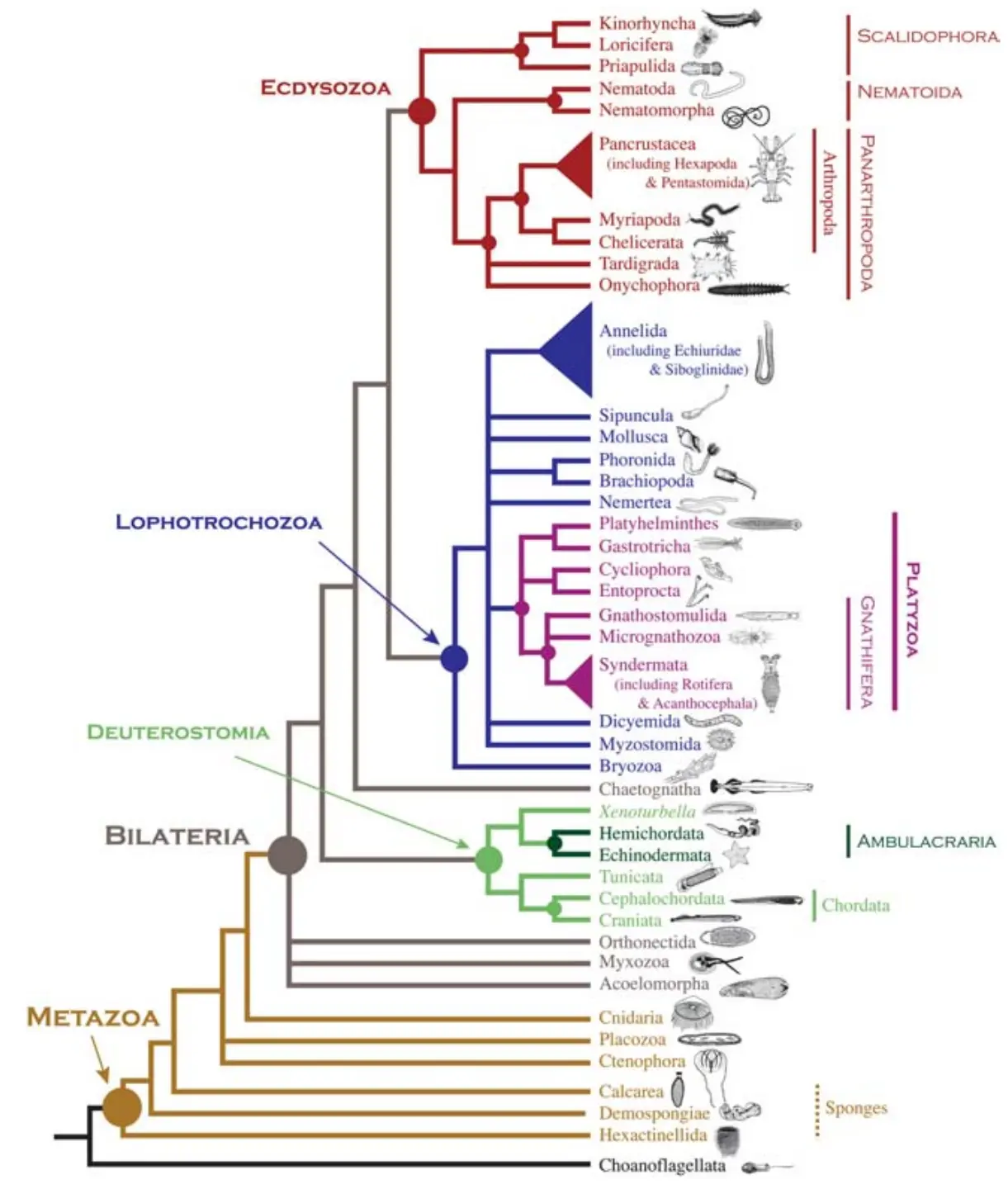
Trees are everywhere in biology…
… an ARG (recombinant genealogy) is a generalisation of an evolutionary tree
The coalescent with recombination
No recombination
With recombination
To the left, an ARG without recombination. To the right, recombination splits lineage 1 going backwards in time, where L_1 takes the left path, and R_1 the right. Consequently, L_1 and R_1 have different MRCAs!
ARG visualization
https://github.com/kitchensjn/tskit_arg_visualizer Wong et al. (2024)
Why ARGs?
Generalises an evolutionary “tree” to genome-wide inheritance
Understanding
- Describes the processes that generated genomic data
- Helps to generate hypotheses
Power
- “True” genealogy represents total DNA history
Size & Speed
- Tree-like structures make algorithms fast
- Can compress variant data: an “evolutionary encoding”
Representation
- Duality ARG - tree sequence along a genome, where a tree is defined over a non-recombining interval
But still early days…
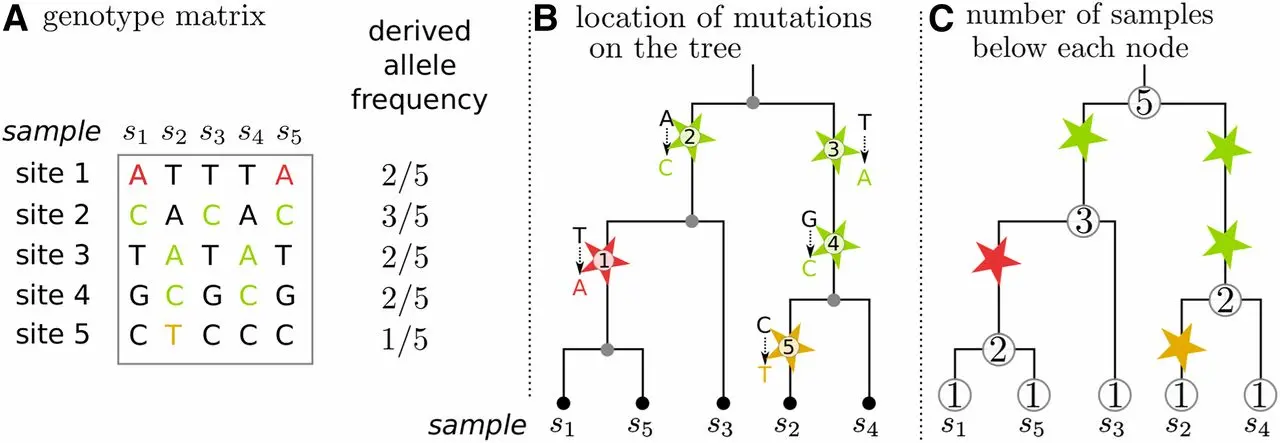
Of genotype matrices and genealogical trees
Efficiently Summarizing Relationships in Large Samples: A General Duality Between Statistics of Genealogies and Genomes. Ralph et al. (2020), Fig. 1
Trees capture biology
Neutral
Expansion
Bottleneck
Selection
msprime stores data as succinct tree sequences
Tree sequences compress data and speedup analyses
- Compact storage (“domain specific compression”)
- Fast, efficient analysis (a “succinct” structure)
- Well tested, open source (active dev community)
Data compression
- Built-in functionality (well documented: http://tskit.dev)
…but limited support for major genomic rearrangements (e.g. inversions, large indels): genomes should be (reasonably) aligned => current primary focus = population genetics
Speed
tskit terminology: the basics
- Multiple local trees exist along a genome of fixed length (by convention measured in base pairs)
- Genomes exist at specific times, and arerepresented by nodes (the same node can persist across many local trees)
- Some nodes are most recent common ancestors (MRCAs) of other nodes
- Entities are zero-based: the rst node has id 0, the second id 1, …
Images from online tutorial “Terminology & concepts” https://tskit.dev/tutorials/terminology_and_concepts.html
tskit terminology: nodes and edges
Nodes (=genomes)
- exist at a specific time
- can be flagged as “samples”
- can belong to “individuals” (e.g., 2 nodes per individuals in humans) and, if useful, “populations”
| id | flags | population | individual | time | metadata |
|---|---|---|---|---|---|
| 0 | 1 | 0 | 0 | 0.00000000 | |
| 1 | 1 | 0 | 0 | 0.00000000 | |
| 2 | 1 | 0 | 1 | 0.00000000 | |
| 3 | 1 | 0 | 1 | 0.00000000 | |
| 4 | 1 | 0 | 2 | 0.00000000 | |
| 5 | 1 | 0 | 2 | 0.00000000 | |
| 6 | 0 | 0 | -1 | 14.70054184 | |
| 7 | 0 | 0 | -1 | 40.95936939 | |
| 8 | 0 | 0 | -1 | 72.52965866 | |
| 9 | 0 | 0 | -1 | 297.22307150 | |
| 10 | 0 | 0 | -1 | 340.15496436 | |
| 11 | 0 | 0 | -1 | 605.35907657 |
Edges
- Connect a parent & child
- Have a left & right genomic coordinate
- Usually span multiple trees (e.g., edges connecting nodes 1+7 and 4+7)
| id | left | right | parent | child | metadata |
|---|---|---|---|---|---|
| 0 | 0 | 1000 | 6 | 2 | |
| 1 | 0 | 1000 | 6 | 5 | |
| 2 | 0 | 1000 | 7 | 1 | |
| 3 | 0 | 1000 | 7 | 4 | |
| 4 | 0 | 1000 | 8 | 3 | |
| 5 | 0 | 1000 | 8 | 6 | |
| 6 | 307 | 1000 | 9 | 0 | |
| 7 | 307 | 1000 | 9 | 7 | |
| 8 | 0 | 307 | 10 | 0 | |
| 9 | 0 | 567 | 10 | 8 | |
| 10 | 307 | 567 | 10 | 9 | |
| 11 | 0 | 307 | 11 | 7 | |
| 12 | 567 | 1000 | 11 | 8 | |
| 13 | 567 | 1000 | 11 | 9 | |
| 14 | 0 | 307 | 11 | 10 |
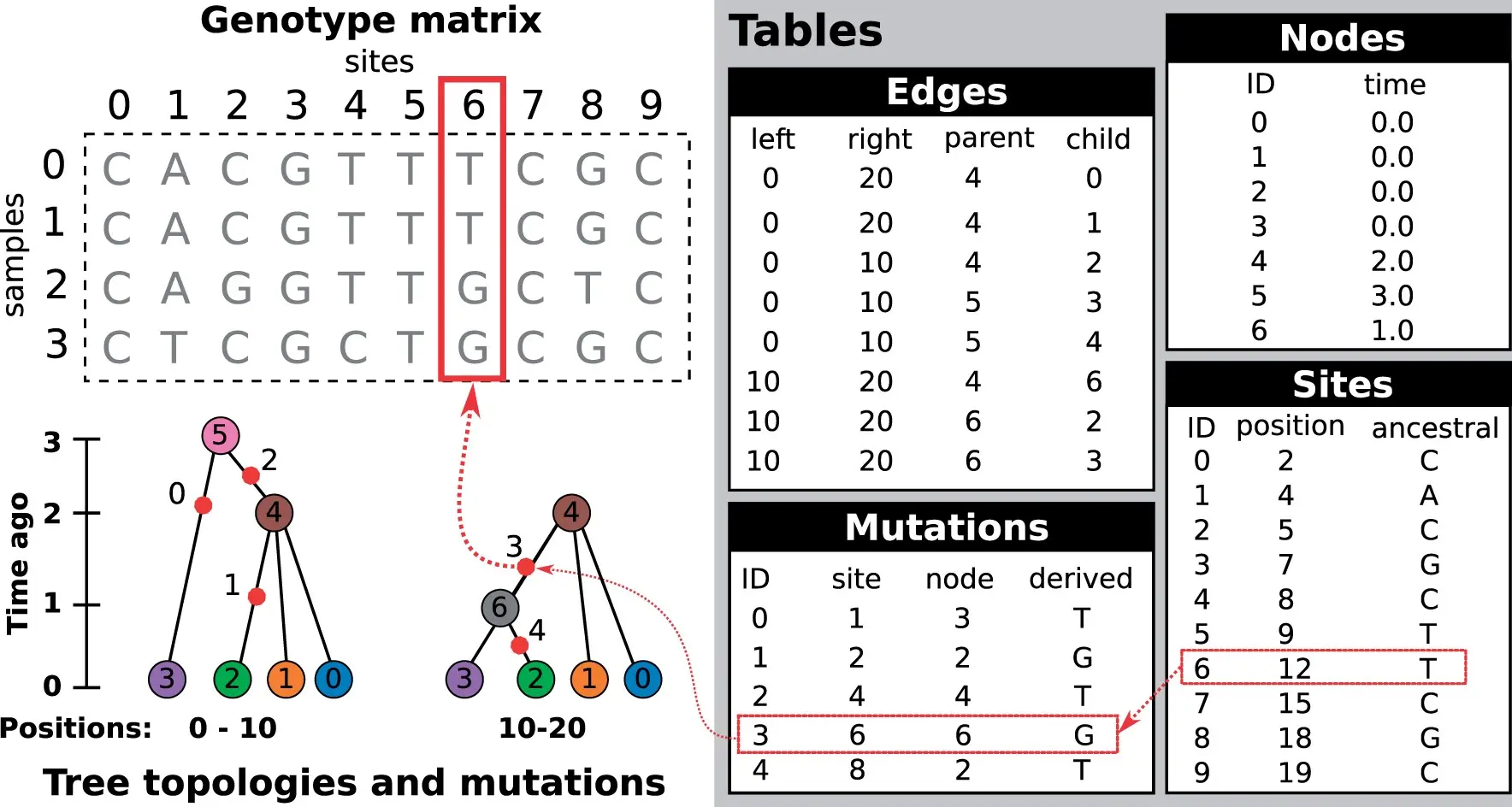
tskit terminology: sites and mutations
This is how we can encode genetic variation. Most genomic positions do not vary between genomes: usually we don’t bother tracking these.
tskit terminology: sites and mutations
We can create a site at a given genomic position with a fixed ancestral state.
| id | position | ancestral_state | metadata |
|---|---|---|---|
| 0 | 52 | C | |
| 1 | 200 | A | |
| 2 | 335 | A | |
| 3 | 354 | A | |
| 4 | 474 | G | |
| 5 | 523 | A | |
| 6 | 774 | C | |
| 7 | 796 | C | |
| 8 | 957 | A |
This is how we can encode genetic variation. Most genomic positions do not vary between genomes: usually we don’t bother tracking these.
tskit terminology: sites and mutations
We can create a site at a given genomic position with a fixed ancestral state.
| id | position | ancestral_state | metadata |
|---|---|---|---|
| 0 | 52 | C | |
| 1 | 200 | A | |
| 2 | 335 | A | |
| 3 | 354 | A | |
| 4 | 474 | G | |
| 5 | 523 | A | |
| 6 | 774 | C | |
| 7 | 796 | C | |
| 8 | 957 | A |
This is how we can encode genetic variation. Most genomic positions do not vary between genomes: usually we don’t bother tracking these.
Normally, a site is created in order to place one or more mutations at that site
| id | site | node | time | derived_state | parent | metadata |
|---|---|---|---|---|---|---|
| 0 | 0 | 8 | 247.85988972 | T | -1 | |
| 1 | 1 | 0 | 169.80687857 | C | -1 | |
| 2 | 2 | 3 | 31.84262397 | C | -1 | |
| 3 | 3 | 9 | 326.26095349 | C | -1 | |
| 4 | 3 | 7 | 71.04212649 | T | 3 | |
| 5 | 4 | 3 | 42.72352948 | C | -1 | |
| 6 | 5 | 7 | 55.44045835 | T | -1 | |
| 7 | 6 | 0 | 259.82567754 | T | -1 | |
| 8 | 7 | 8 | 169.87040769 | G | -1 | |
| 9 | 8 | 0 | 42.47396523 | C | -1 |
Tree sequence inference methods
tsinfer/tsdate relate ARGweaver SINGER ARGNeedle Threads
Analysis of tree sequences with 
Adapted from slide by Yun Deng.
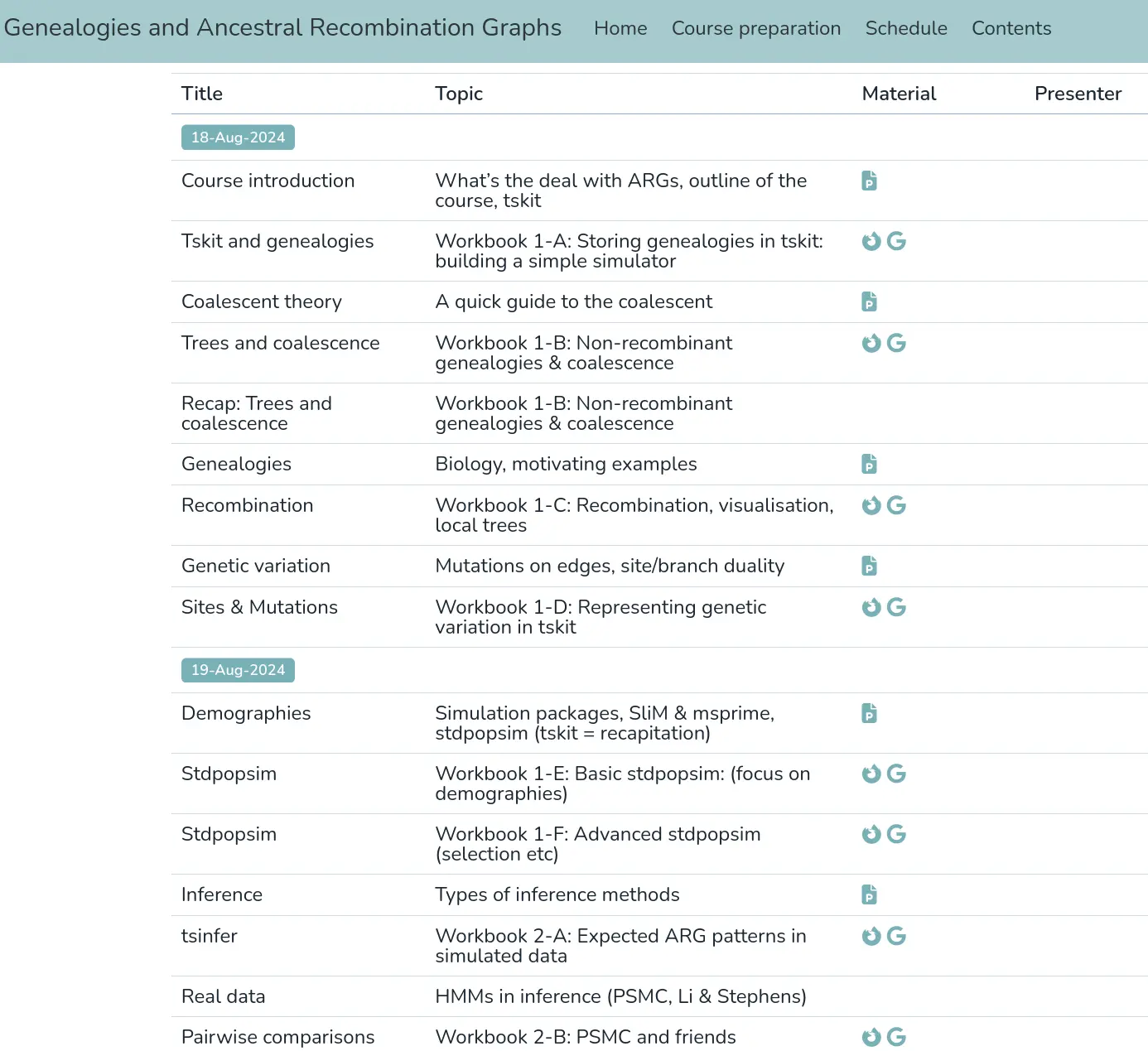
Relate
- fast, but limited sample sizes (1000s?)
- good support for ancient DNA
tsinfer - tree sequence inference
- fast
- scales! (millions of samples!)
- introduces tree sequence format
- only genealogies, no branch lengths (but see tsdate (Wohns et al., 2022)
GARG workshop Drøbak research station Aug-24
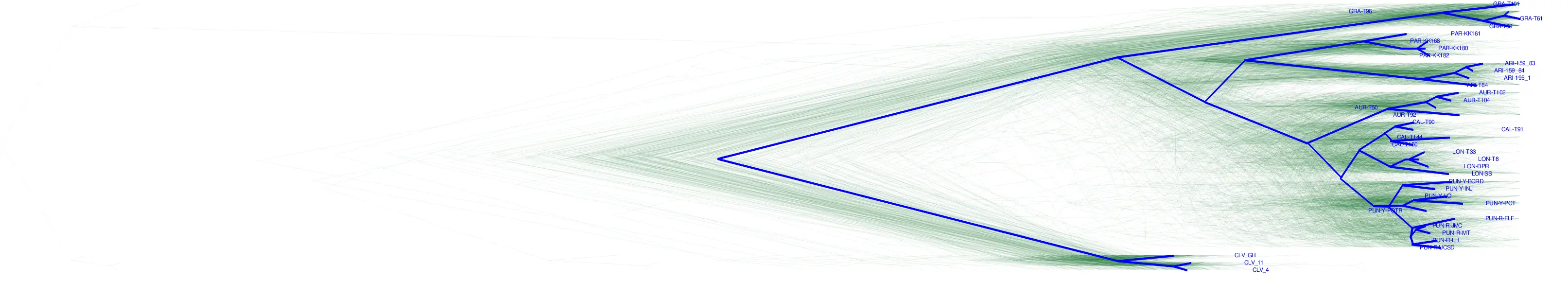
Mapping workflow Monkeyflower
Tree sequence inference in Monkeyflower

Bibliography
Population Genomics in Practice