Population genomics in practice
What is population genomics?
Intended learning outcomes
Course
- Present minimum toolkit of methods that should be known to anyone starting out in population genomics
- Sufficiently small for one-week workshop
Lecture
- Present practical example of toolkit as applied in (Fuller et al., 2020)
- Briefly discuss baseline model (Johri et al., 2022)
Example: Population genetics of the coral Acropora millepora
Motivation: corals are facing hard times and to prevent future losses of coral cover a better understanding of genetics is warranted.
Genome assembly and sampling
Motivation: most analyses require a reference sequence with which to compare resequenced samples
- Assemble high-quality reference genome
- Choice of populations, sampling locations
Fuller et al. (2020)
Example: Population genetics of the coral Acropora millepora
Motivation: corals are facing hard times and to prevent future losses of coral cover a better understanding of genetics is warranted.
Describe genetic structure and demographic history
Motivation:
- address basic question of why genetic structure looks the way it does
- demographic history may generate signals similar to selection
Fuller et al. (2020)
Example: Population genetics of the coral Acropora millepora
Motivation: corals are facing hard times and to prevent future losses of coral cover a better understanding of genetics is warranted.
Characterize population structure
Motivation:
- identify populations for contrasts in e.g. selection scans
- identify admixed individuals that should be removed from analyses
- identify barriers to gene flow etc
Fuller et al. (2020)
Example: Population genetics of the coral Acropora millepora
Motivation: corals are facing hard times and to prevent future losses of coral cover a better understanding of genetics is warranted.
Genomic scans for selection
Motivation: identify loci associated with adaptation / selection
- little differentiation over reefs, however thermal regimes
- genomic scan for \pi (diversity) outliers
Fuller et al. (2020)
Example: Population genetics of the coral Acropora millepora
Study highlights common analyses in population genomics study:
- Genome assembly, resequencing, variant calling and filtering
- Description of variation (e.g., \pi) and genetic structure (LD)
- Description of population structure (admixture, PCA)
- Modelling of demographic history (PSMC)
- Genome scans for adaptive traits
Population genetics
From population genetics to population genomics
First study of natural population. However, limited to one locus.
From population genetics to population genomics
Same system but genome-wide. Plots represent all chromosomes and the entire genome.
From population genetics to population genomics
Novelty: now possible to do genome-wide characterization of variation in different functional contexts
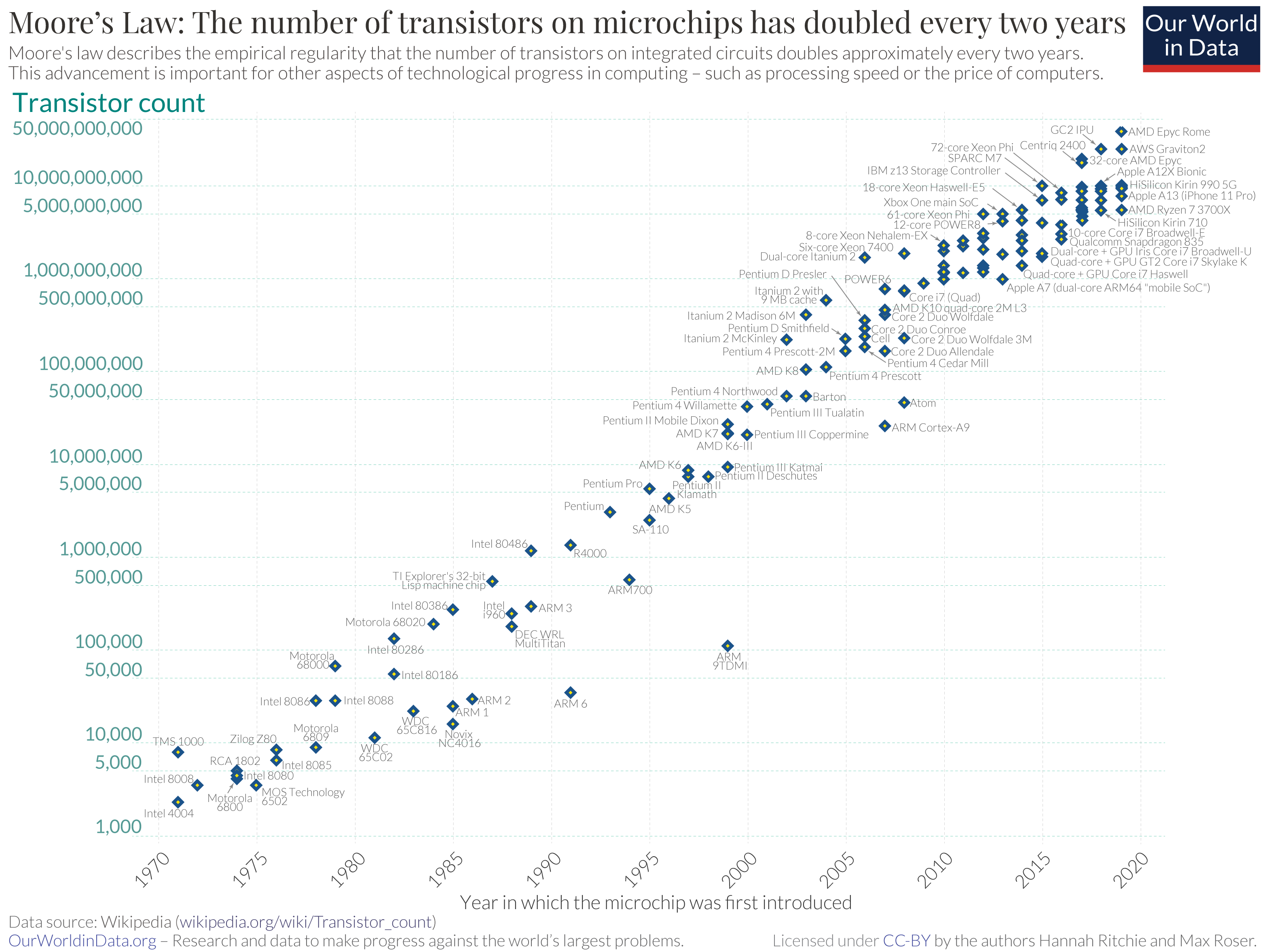
The technological revolution in sequencing and computing
Statistical inference in population genomics
The data deluge requires advanced statistical methods and models to do inference. Today data production outpaces theoretical advances. Therefore, take care not to attach too much faith to a test that explains data well.
A population genomics study should aim at generating a baseline model that takes into account the processes that shape genetic variation (Johri et al., 2022):
- mutation
- recombination
- gene conversion
- purifying selection acting on functional regions and its effects on linked variants (background selection)
- genetic drift with demographic history and geographic structure
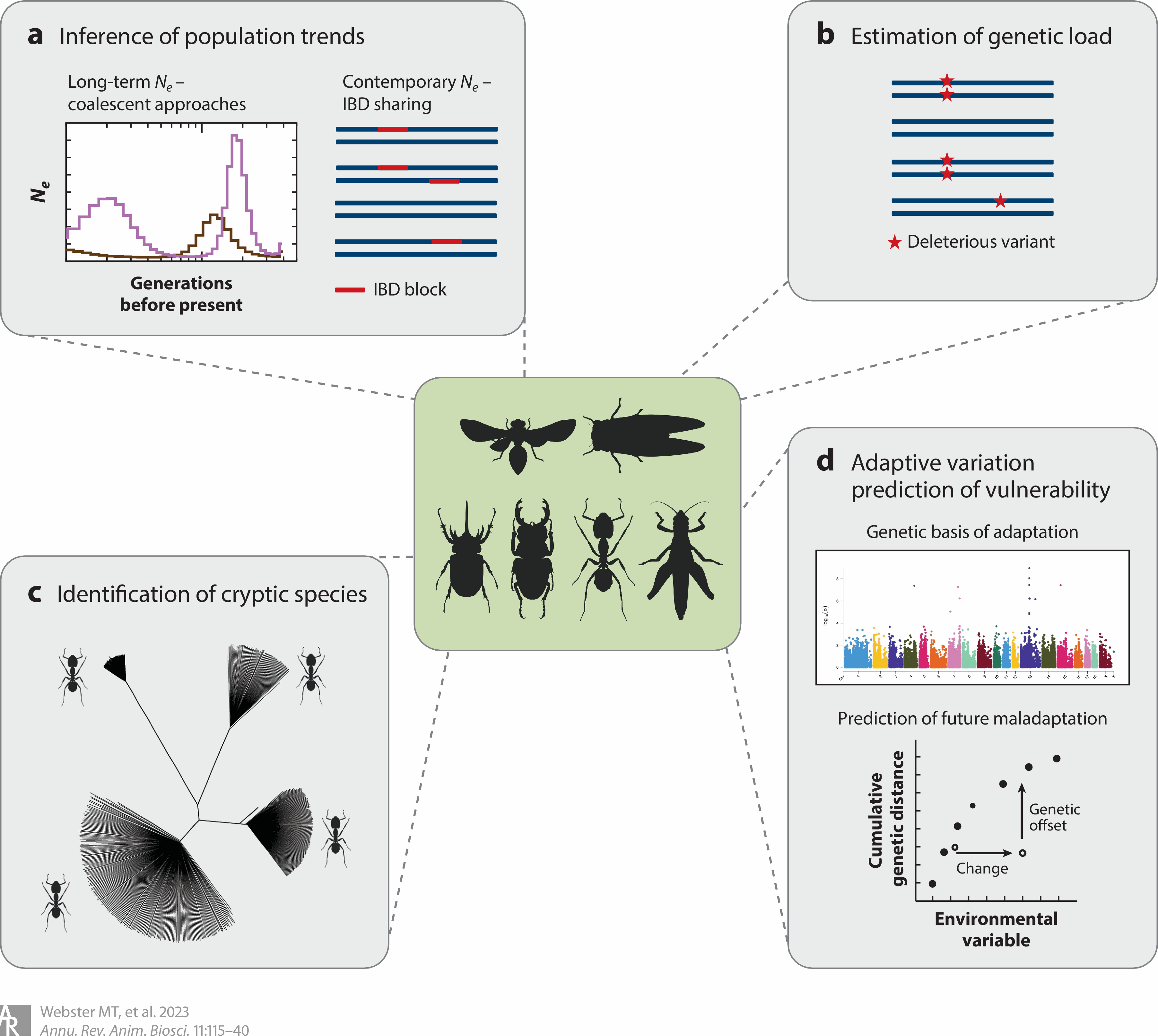
Applications of population genomics
Bibliography
Population Genomics in Practice