The Wright-Fisher model
Of population models and genealogies
Models of populations
Wright-Fisher model
Model of populations that describes genealogical relationships of genes (chromosomes) in a population under the following assumptions (Hein et al., 2005):
- discrete and non-overlapping generations
- haploid individuals or two subpopulations (males and females)
- constant population size
- all individuals are equally fit
- population has no geographical or social structure
- no recombination
Wright-Fisher model
Model of populations that describes genealogical relationships of genes (chromosomes) in a population under the following assumptions (Hein et al., 2005):
- discrete and non-overlapping generations
- haploid individuals or two subpopulations (males and females)
- constant population size
- all individuals are equally fit
- population has no geographical or social structure
- no recombination
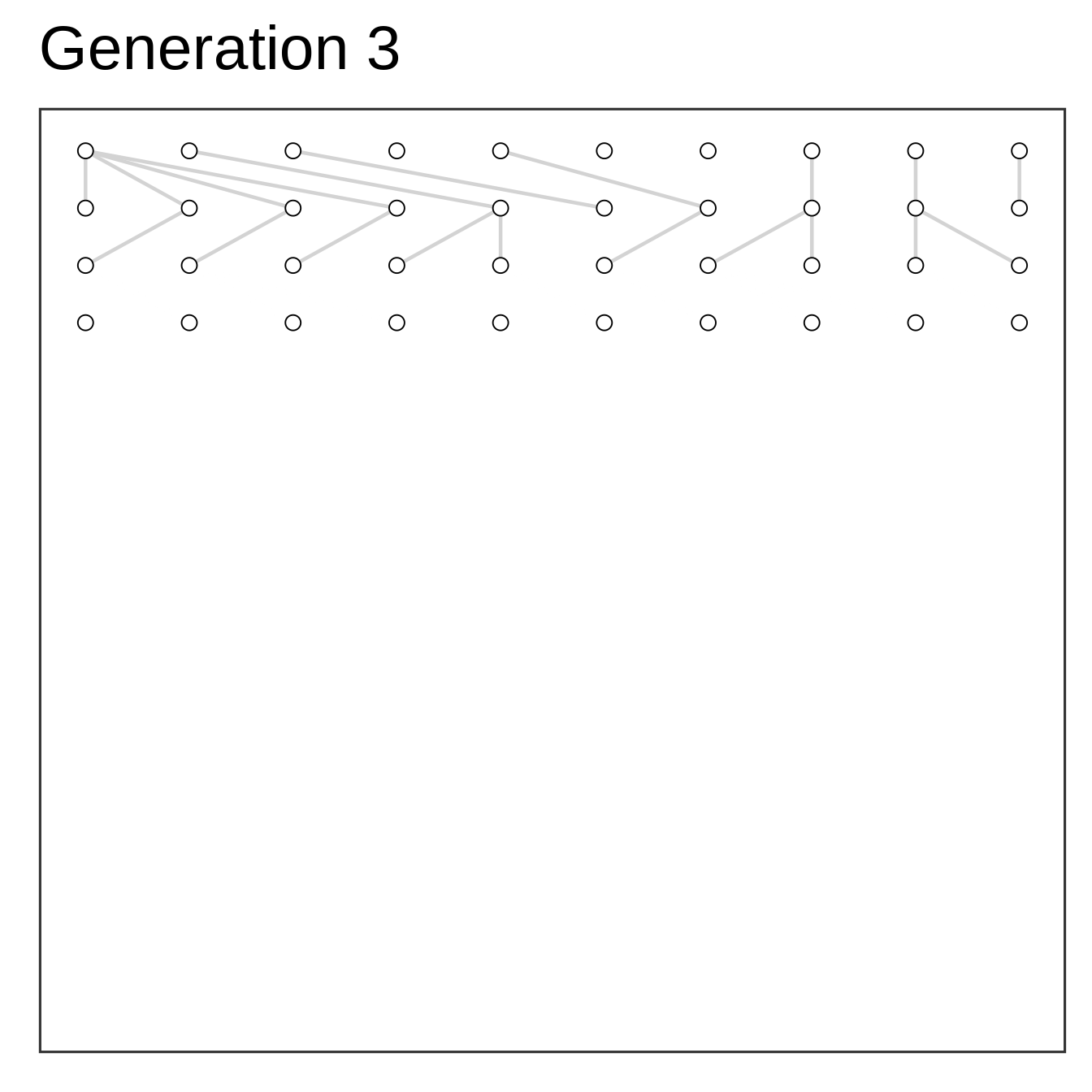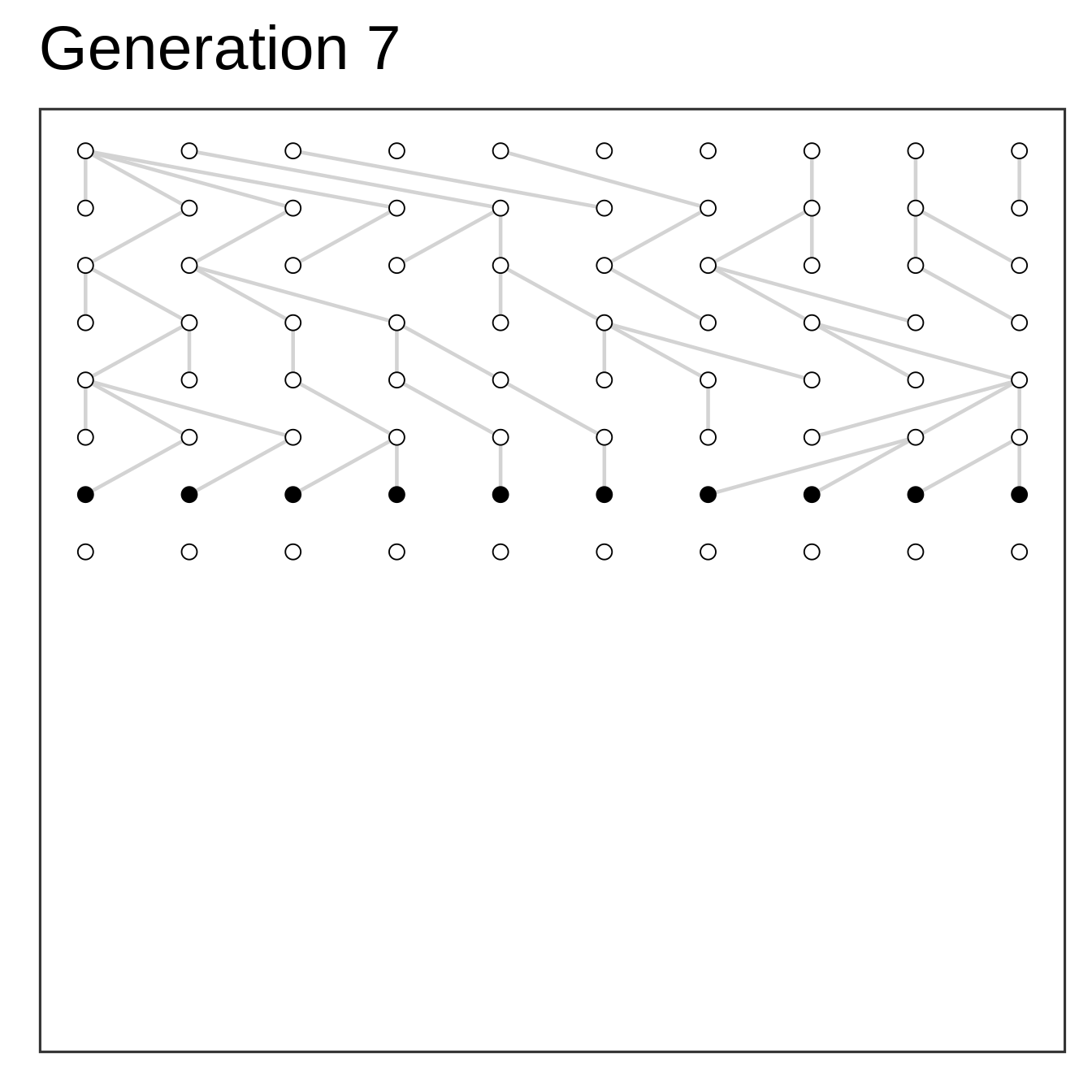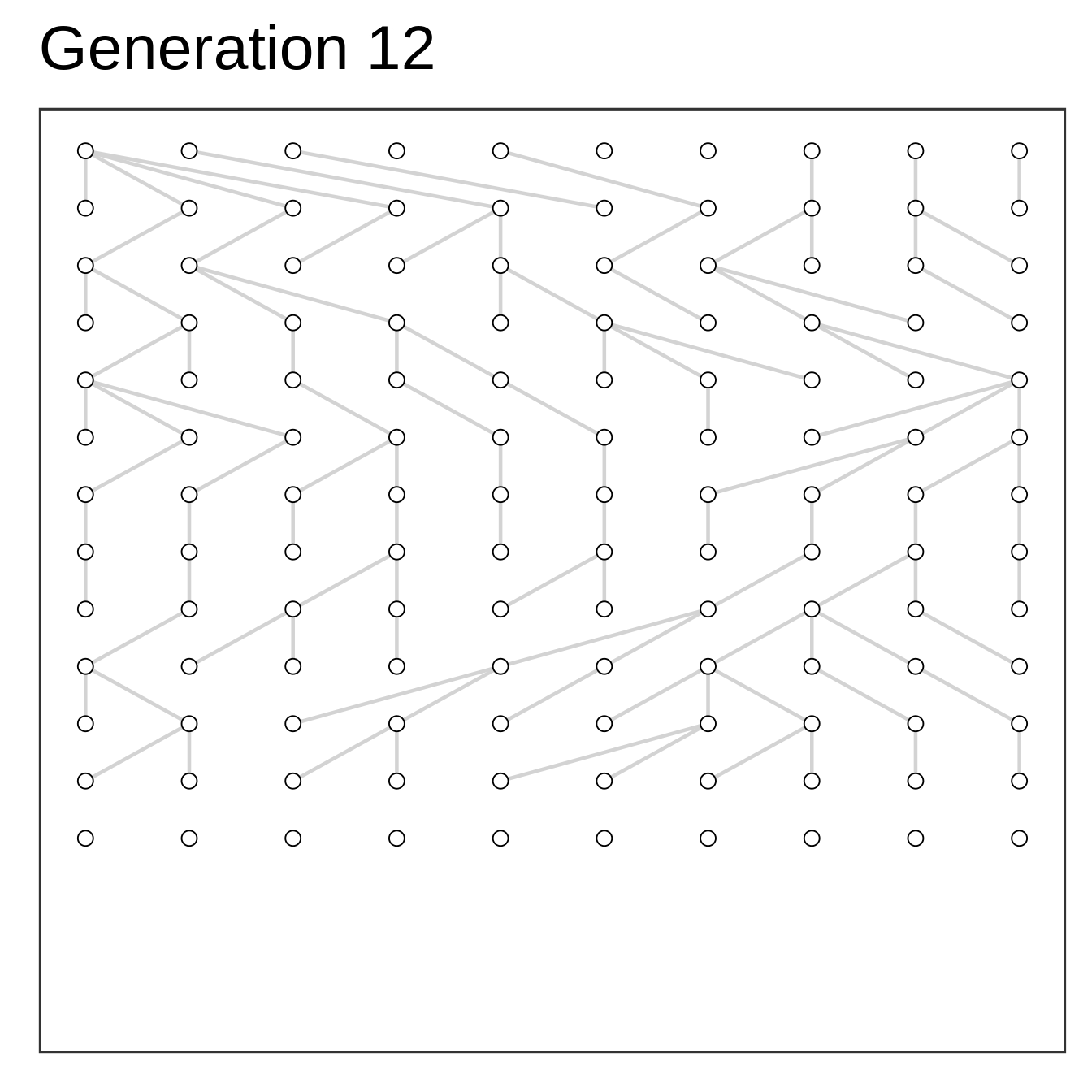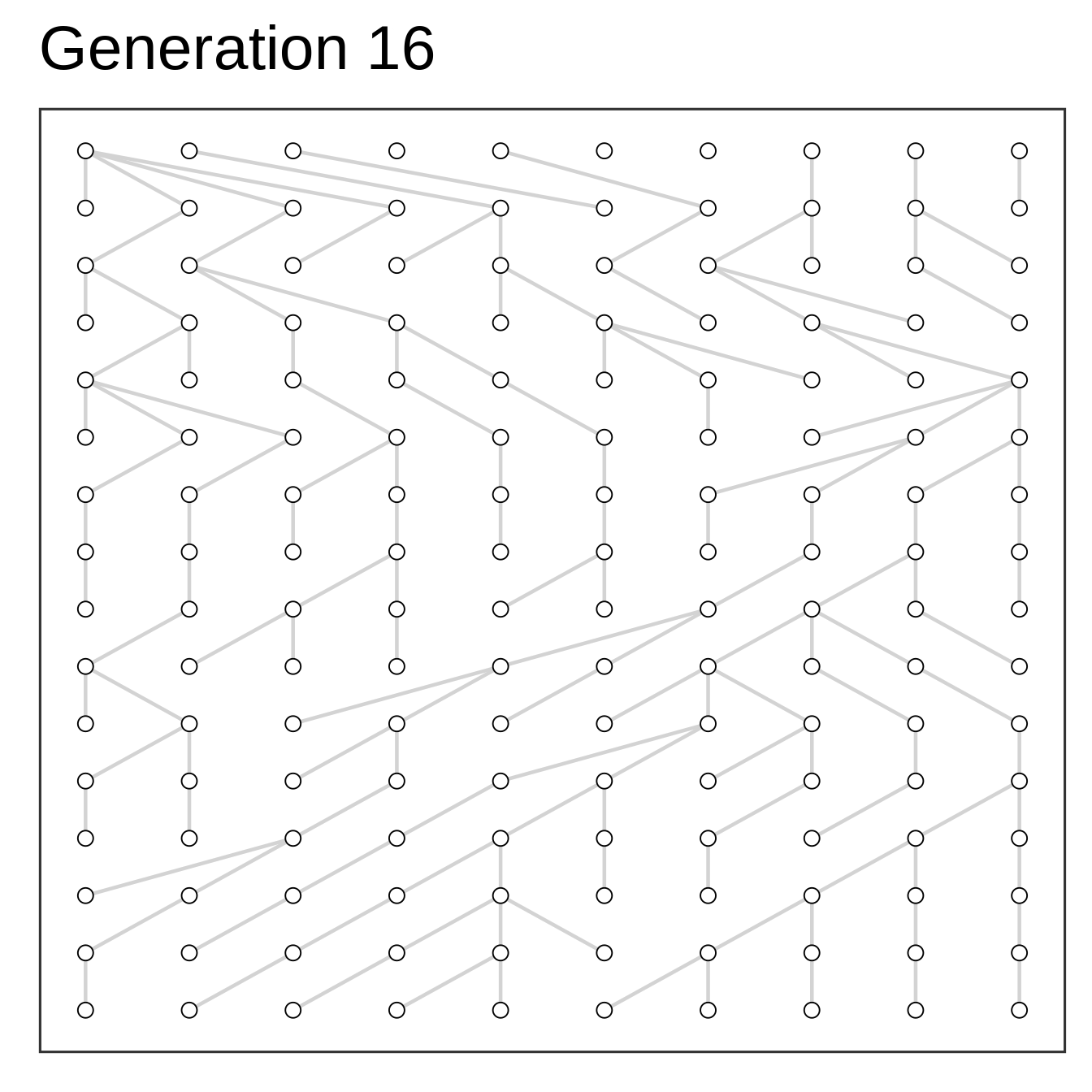
Algorithm
- Setup starting population at time zero
Wright-Fisher model
Model of populations that describes genealogical relationships of genes (chromosomes) in a population under the following assumptions (Hein et al., 2005):
- discrete and non-overlapping generations
- haploid individuals or two subpopulations (males and females)
- constant population size
- all individuals are equally fit
- population has no geographical or social structure
- no recombination
Algorithm
- Setup starting population at time zero
- Add offspring (same size) at time one
Wright-Fisher model
Model of populations that describes genealogical relationships of genes (chromosomes) in a population under the following assumptions (Hein et al., 2005):
- discrete and non-overlapping generations
- haploid individuals or two subpopulations (males and females)
- constant population size
- all individuals are equally fit
- population has no geographical or social structure
- no recombination
Algorithm
- Setup starting population at time zero
- Add offspring (same size) at time one
- Select parents to offspring at random
Wright-Fisher model
Model of populations that describes genealogical relationships of genes (chromosomes) in a population under the following assumptions (Hein et al., 2005):
- discrete and non-overlapping generations
- haploid individuals or two subpopulations (males and females)
- constant population size
- all individuals are equally fit
- population has no geographical or social structure
- no recombination
Algorithm
- Setup starting population at time zero
- Add offspring (same size) at time one
- Select parents to offspring at random
Wright-Fisher model
Model of populations that describes genealogical relationships of genes (chromosomes) in a population under the following assumptions (Hein et al., 2005):
- discrete and non-overlapping generations
- haploid individuals or two subpopulations (males and females)
- constant population size
- all individuals are equally fit
- population has no geographical or social structure
- no recombination
Algorithm
- Setup starting population at time zero
- Add offspring (same size) at time one
- Select parents to offspring at random
Wright-Fisher model
Model of populations that describes genealogical relationships of genes (chromosomes) in a population under the following assumptions (Hein et al., 2005):
- discrete and non-overlapping generations
- haploid individuals or two subpopulations (males and females)
- constant population size
- all individuals are equally fit
- population has no geographical or social structure
- no recombination
Algorithm
- Setup starting population at time zero
- Add offspring (same size) at time one
- Select parents to offspring at random
Wright-Fisher model
Model of populations that describes genealogical relationships of genes (chromosomes) in a population under the following assumptions (Hein et al., 2005):
- discrete and non-overlapping generations
- haploid individuals or two subpopulations (males and females)
- constant population size
- all individuals are equally fit
- population has no geographical or social structure
- no recombination
Algorithm
- Setup starting population at time zero
- Add offspring (same size) at time one
- Select parents to offspring at random
Wright-Fisher model
Model of populations that describes genealogical relationships of genes (chromosomes) in a population under the following assumptions (Hein et al., 2005):
- discrete and non-overlapping generations
- haploid individuals or two subpopulations (males and females)
- constant population size
- all individuals are equally fit
- population has no geographical or social structure
- no recombination
Algorithm
- Setup starting population at time zero
- Add offspring (same size) at time one
- Select parents to offspring at random
Wright-Fisher model
Model of populations that describes genealogical relationships of genes (chromosomes) in a population under the following assumptions (Hein et al., 2005):
- discrete and non-overlapping generations
- haploid individuals or two subpopulations (males and females)
- constant population size
- all individuals are equally fit
- population has no geographical or social structure
- no recombination
Algorithm
- Setup starting population at time zero
- Add offspring (same size) at time one
- Select parents to offspring at random
Wright-Fisher model
Wright-Fisher model
The Wright-Fisher sampling model
Let’s formalise the sampling process of the Wright-Fisher model1. We assume
- a single locus in a haploid population of size 2N (or diploid of size N when random mating)
- no mutation and selection
- discrete generations
Each generation we sample 2N new chromosomes from the previous generation. The probability of choosing a chromosome v is 1/2N (coin flip with probability of success 1/2N). Since the trials are independent, and we perform 2N trials, the number of offspring k of a given chromosome v is binomially distributed \mathrm{Bin}(m, p), with parameters m=2N and probability of success p=\frac{1}{2N}.
Properties of Wright-Fisher sampling
Bibliography
Wright-Fisher model