Genetic diversity
The origin and maintenance of genetic diversity
Origin and change of variation
Wright-Fisher model with alleles
Binomial process models allele sampling
We assume two alleles A, a, each with i and j=2N-i copies in generation t.
i=8, j=2\cdot 6-8=4
Let p_t=i/2N be the frequency of A in generation t, and q_t=1-p_t the frequency of a.
p_t = 8/12
p_{t+1} = 4/12
Prob(k A alleles in next generation) is \mathsf{Bin}(2N, \frac{i}{2N})
Genetic drift
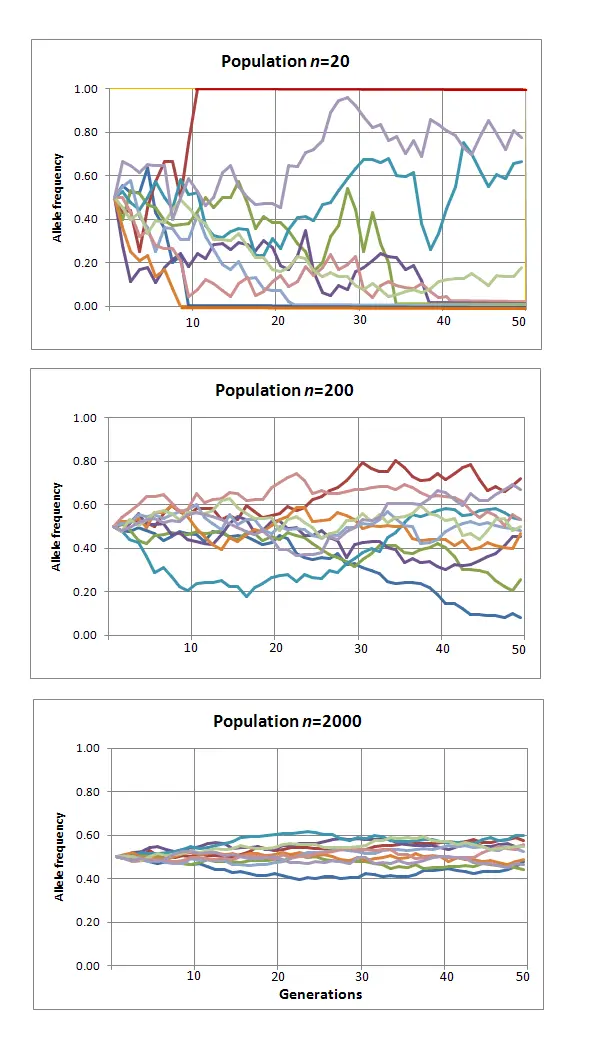
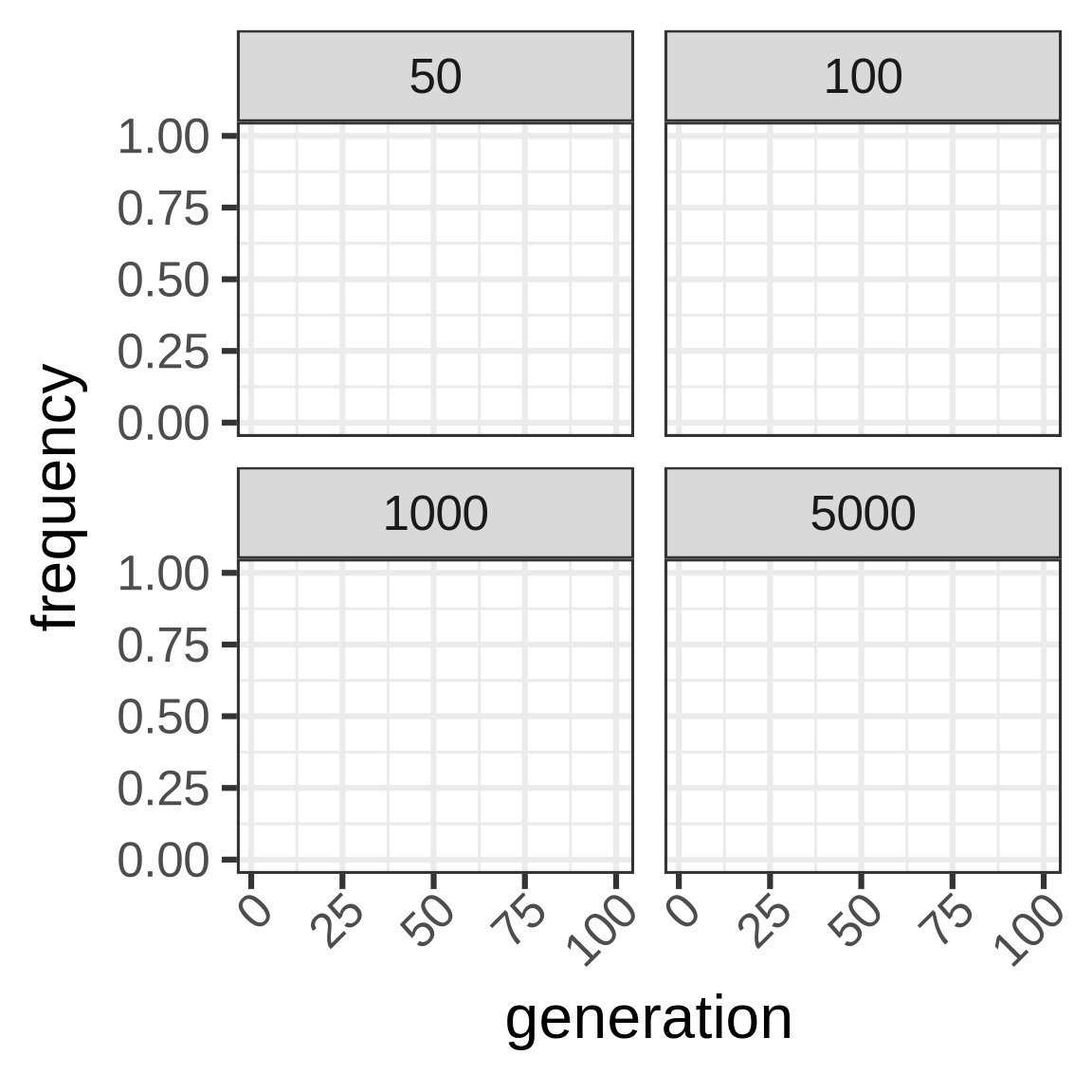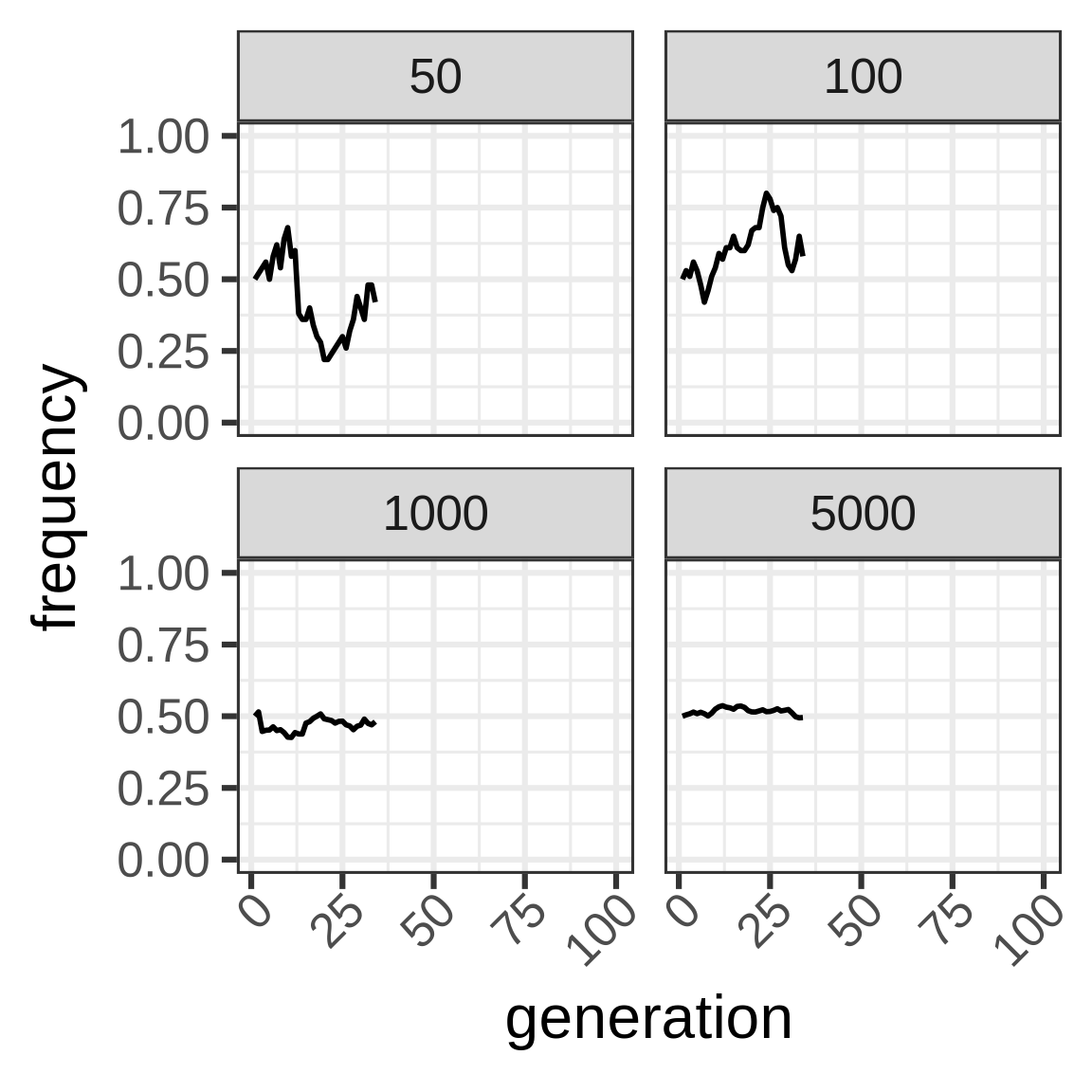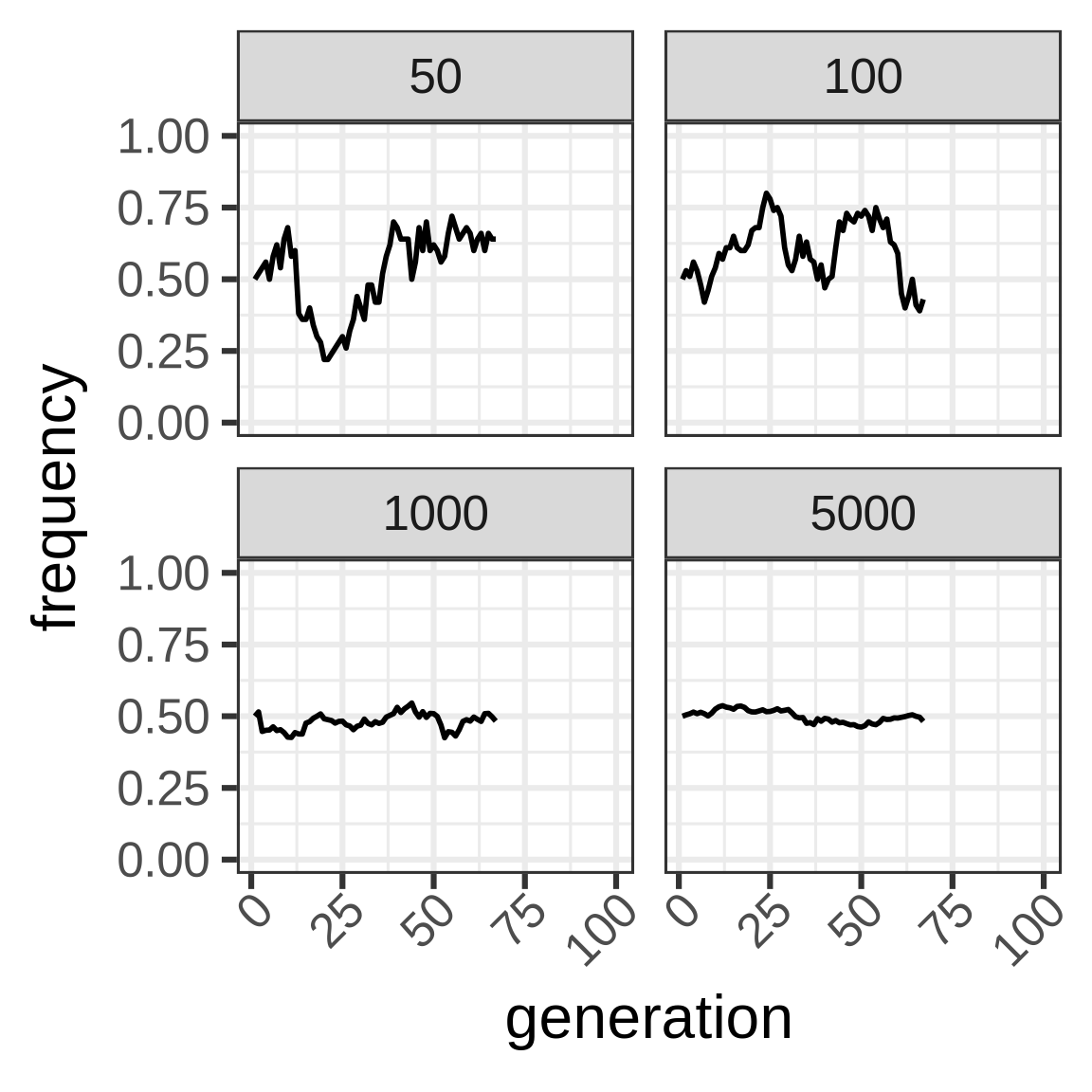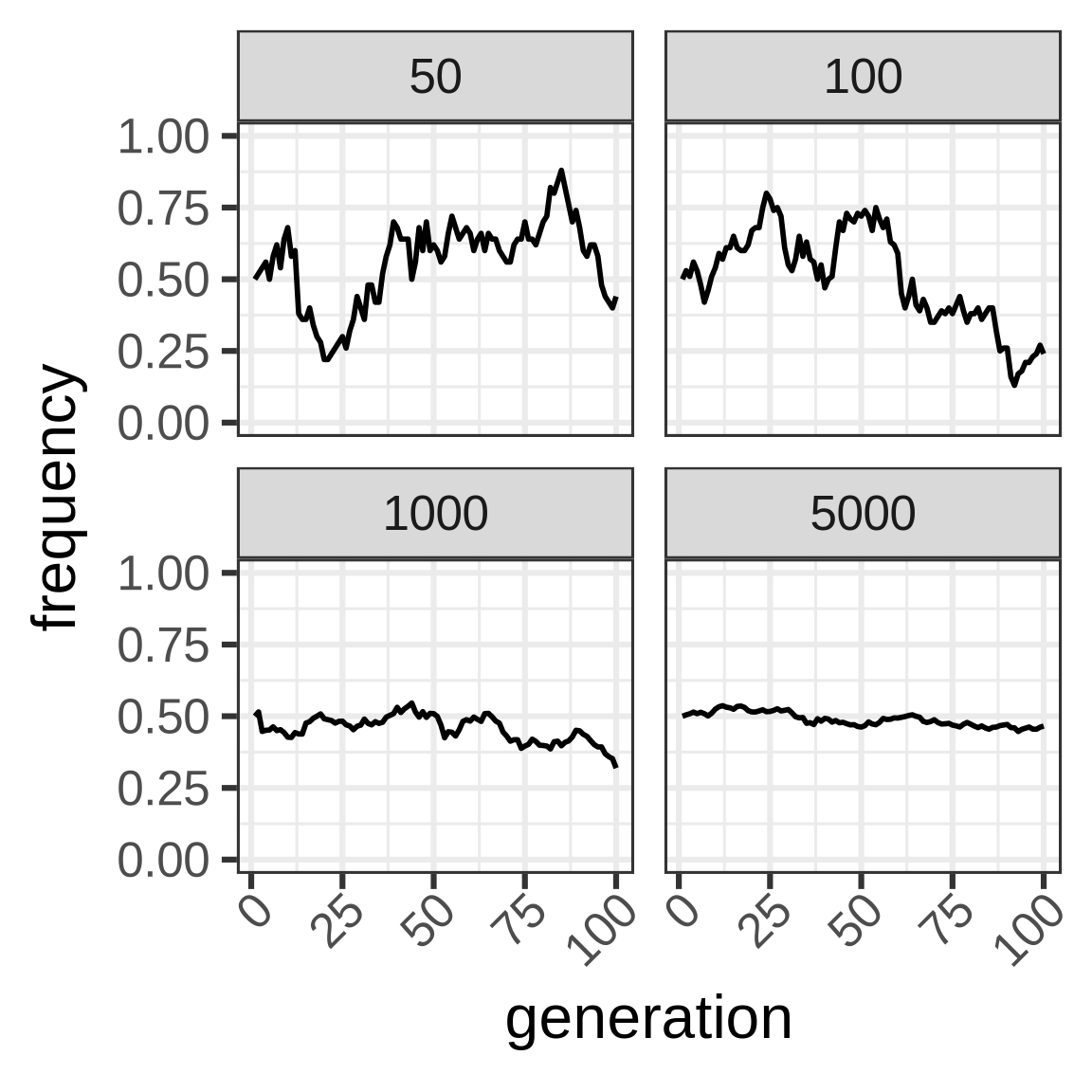
To capture dynamics, follow allele frequency trajectory (p_t) as function of time.
##' Wright Fisher model - follow allele frequency distribution
##'
##' @param p0 Starting frequency
##' @param n Population size
##' @param generations Number of generations to simulate
##'
wright_fisher <- function(p0, n, generations) {
x <- vector(mode = "numeric", length = generations)
x[1] <- p0
for (i in seq(2, length(x))) {
x[i] <- rbinom(1, size = n, prob = x[i - 1])/n
}
x
}Genetic drift
Allele frequency distribution for N=1
Instead of looking at frequencies let’s switch to distributions of alleles for one individual, one locus. Then there are three possible genotypes (states) aa, aA, and AA. Let n=0,1,2 be an integer corresponding to each genotype (i.e., it counts the number of A alleles).
Assume individual mates with itself at random(!) starting in either of the three states. How does distribution evolve?
Probability distributions of allele frequencies
Mathematical treatment of drift can become complicated: easier to study dynamics of heterozygosity
Heterozygosity dynamics
Let \mathcal{H}_t be the probability that two alleles are different by state. One can show that the time course evolution of \mathcal{H}_t in a randomly mating population consisting of N diploid hermaphroditic individuals is
\mathcal{H}_t = \mathcal{H}_0 \left( 1 - \frac{1}{2N} \right)^t
Important consequence: heterozygosity in WF population lost at rate 1/2N.
Heterozygosity dynamics
Effective population size
Assumptions underlying Wright-Fisher model seldom fulfilled for natural populations. In particular
- non-random mating (population structure)
- fluctuations of population census size
Therefore, magnitude of drift experienced by a population different from that predicted by population size
Technically correct definition (but see Waples (2022), Waples (2025)):
N_e is the size of an ideal population that would experience the same rate of genetic drift as the population in question.
Mutation
Mutation and drift
Genetic drift “moves” frequencies to the point that variation is lost via allele fixation or loss. New variation is introduced through mutation. We typically assume mutations are described by a Poisson process with rate \mu (per generation).
The mutation rate is denoted \mu, and the population scaled mutation rate is 2N_e\mu for haploid populations, 4N_e\mu for diploid, where N_e is the effective population size.
The mutation - drift balance is when the diversity lost due to drift equals the diversity gained due to mutation.
Tracing the evolution of mutations
Mutation drift balance
Drift removes variation. Mutation reintroduces it. At equilibrium the change in variation by definition is 0. In terms of \mathcal{H}_t (the probability that two alleles are not identical by state), \Delta\mathcal{H}=0.
One can show1 the classical formula that the equilibrium heterozygosity value is
\hat{\mathcal{H}} = \frac{4N_e\mu}{1 + 4N_e\mu}
\mu is often assumed known, and heterozygosity is easily calculated from data, which provides a way of estimating N_e.
The compound parameter 4N_e\mu is called the population scaled mutation rate and is commonly named \theta such that
\hat{\mathcal{H}} = \frac{\theta}{1 + \theta}
The neutral theory of evolution
Mutation drift balance, together with the observation during 50’s-60’s that polymorphism was more common than expected, is the foundation of the neutral theory of evolution (Kimura, 1983): allele frequencies may change and fix due to chance alone and not selection; most mutations behave as if they are neutral.
Nearly neutral theory (Ohta, 1973) was later developed to explain failure to predict scaling of polymorphism with population size: most mutations are not neutral but slightly deleterious and purged from population by natural selection.
Mutation rate can be estimated from substitution rate
Mutation enters populations and may be fixed by drift. Therefore, with time there will be fixed differences, or substitions (typically in the evolution of species) between populations, or species. In molecular evolution, the substition rate, \rho, is the most interesting quantity.
The total number of new mutations in every generation is 2N\mu (total number of gametes times mutation rate)
New mutations fix at a rate 1/2N
Therefore, the average rate of substitution, \rho, is 2N\mu\times1/2N, or
\rho=\mu
which is independent of population size!
Practical implication: we can estimate mutation rate from the substitution rate at neutrally evolving sites (e.g., Kumar & Subramanian (2002))
Bibliography
Genetic diversity