Alleles and genealogies
More descriptions of genetic data
Alleles as algebraic entities
Recall: alleles refer to different variants of a sequence at a locus (genomic position).
Whatever the underlying molecular nature (gene, chromosome, nucleotide, protein), let’s represent a locus by a letter, e.g., A (B if two loci, and so on)
If locus has many alleles 1, 2, ... , could use indexing A_1, A_2, ....
Will use combination A, a for bi-allelic loci from now on
Example: gene coding for flower color
Alleles and frequencies
We will be interested in looking at the dynamics of alleles, i.e., how their abundances in the population change over time. Therefore we want to measure the frequencies of alleles A and a.
Example
Assume following population (n=10, with n_{AA}=5, n_{Aa}=4, n_{aa}=1):









Let p be frequency of A alleles, q=1-p frequency of a alleles; then
5 AA individuals, 4 Aa individuals \Rightarrow p=\frac{5\cdot2 + 4\cdot1}{10\cdot2}=\frac{14}{20}=0.7
and q=1-p=\frac{6}{20}=0.3
Inserting frequencies into Punnett square gives expected frequency of offspring genotypes.
In absence of evolutionary forces alleles are in equilibrium
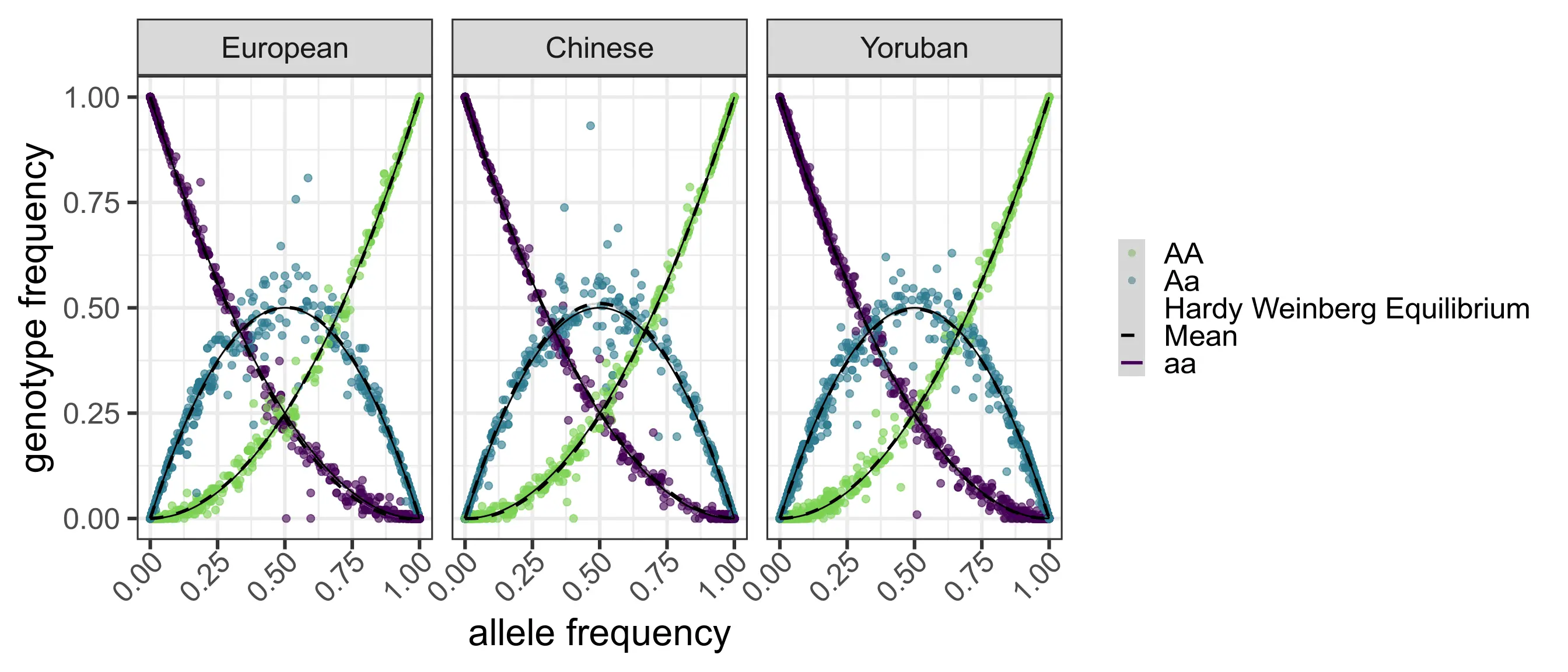
The Hardy-Weinberg equilibrium
For a locus, let A and a be two different alleles and let p be the frequency of the A allele and q=1-p the frequency of the a allele. In the absence of mutation, drift, migration, and other evolutionary processes, the equilibrium state is given by the Hardy-Weinberg equilibrium (HWE).
| A (p) | a (q) | |
|---|---|---|
| A (p) | p^2 | pq |
| a (q) | qp | q^2 |
| Genotype: | AA | Aa | aa |
| Frequency: | p^2 | 2pq | q^2 |
| f_{AA} | f_{Aa} | f_{aa} |
HWE assumption
Under HWE assumption, neither allele nor genotype frequencies change over time.
Importantly, we can calculate allele frequencies from genotype frequencies and vice versa.
p = f_{AA} + \frac{f_{Aa}}{2} = p^2 + pq\\ q = f_{aa} + \frac{f_{Aa}}{2} = q^2 + pq\\
Natural populations do mate randomly?
The Wahlund effect and population substructure
Population P1




p_A = 1 \Rightarrow p_A^2 = 1, p_a^2=2p_Ap_a=0
Population P1




p_a = 1 \Rightarrow p_a^2 = 1, p_A^2=2p_Ap_a=0
Both subpopulations are in HWE!
Population P1+P2:
p_A=p_a=0.5 so we would expect 50% heterozygotes - but there are none!
This is known as the Wahlund effect where the loss of heterozygosity is due to population substructure.
Summarising allele frequencies
Going back to the DNA example let’s tabulate the minor allele frequencies (MAFs):
| 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9 | 10 | 11 | 12 | 13 | 14 | 15 | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| T | T | A | C | A | A | T | C | C | G | A | T | C | G | T | |
| T | T | A | C | G | A | T | G | C | G | C | T | C | G | T | |
| T | C | A | C | A | A | T | G | C | G | A | T | G | G | A | |
| T | T | A | C | G | A | T | G | C | G | C | T | C | G | T | |
| MAF | 0 | 1 | 0 | 0 | 2 | 0 | 0 | 1 | 0 | 0 | 2 | 0 | 1 | 0 | 1 |
The obsession of population genetics
Population genetics is about (Gillespie, 2004)
- describing the genetic structure of populations
- constructing theories on the forces that influence genetic variation
Questions to ponder:
- why does variation look the way it does?
- how is variation maintained?
- how does variation change over time (\Delta p)?
- what forces shape the genetic structure of populations?
p=0.1
\large\rightarrow
p=0.5
\large\rightarrow
p=0.9
Bibliography
Foundations - alleles and genealogies