Trajectory inference analysis: PAGA¶
Partly following tutorial https://scanpy-tutorials.readthedocs.io/en/latest/paga-paul15.html
Loading libraries¶
import numpy as np
import pandas as pd
import matplotlib.pyplot as pl
from matplotlib import rcParams
import scanpy as sc
import scipy
import numpy as np
import matplotlib.pyplot as plt
sc.settings.verbosity = 3 # verbosity: errors (0), warnings (1), info (2), hints (3)
sc.logging.print_versions()
sc.settings.set_figure_params(dpi=100, frameon=False, figsize=(5, 5), facecolor='white', color_map = 'viridis_r')
Loading data¶
In order to speed up the computations during the exercises, we will be using a subset of a bone marrow dataset (originally containing about 100K cells). The bone marrow is the source of adult immune cells, and contains virtually all differentiation stages of cell from the immune system which later circulate in the blood to all other organs.
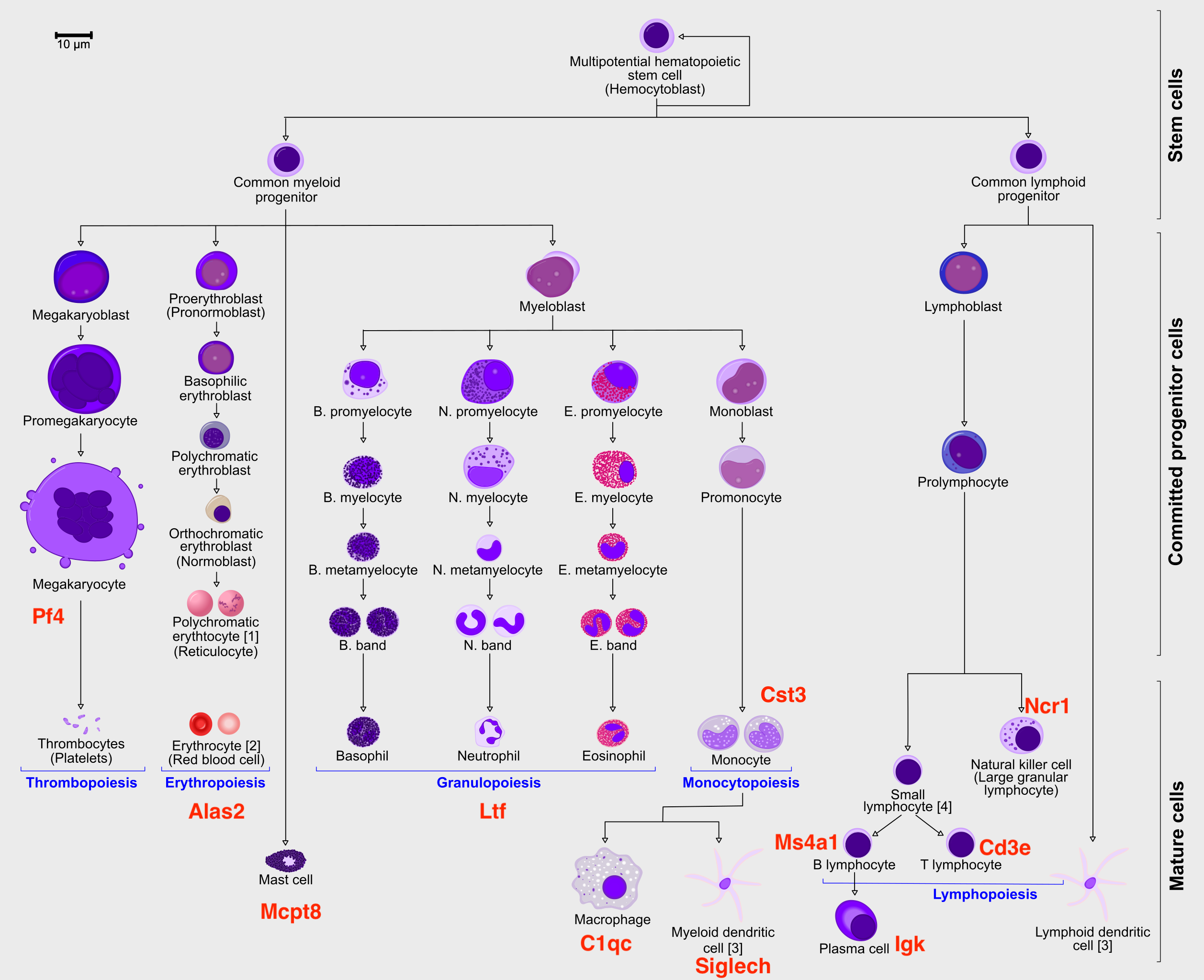
All the data has been preprocessed with Seurat. The file trajectory_scanpy_filtered.h5ad was converted from the Seurat object using the SeuratDisk package. For more information on how it was done, have a look at the script: convert_to_h5ad.R in the github repo.
infile = './../data/bone_marrow/trajectory_scanpy_filtered.h5ad'
adata = sc.read_h5ad(infile)
# check what is in the object
adata
Check that the variable names are correct.
adata.var
# check what you have in the X matrix, should be lognormalized counts.
print(adata.X[:10,:10])
Explore the data¶
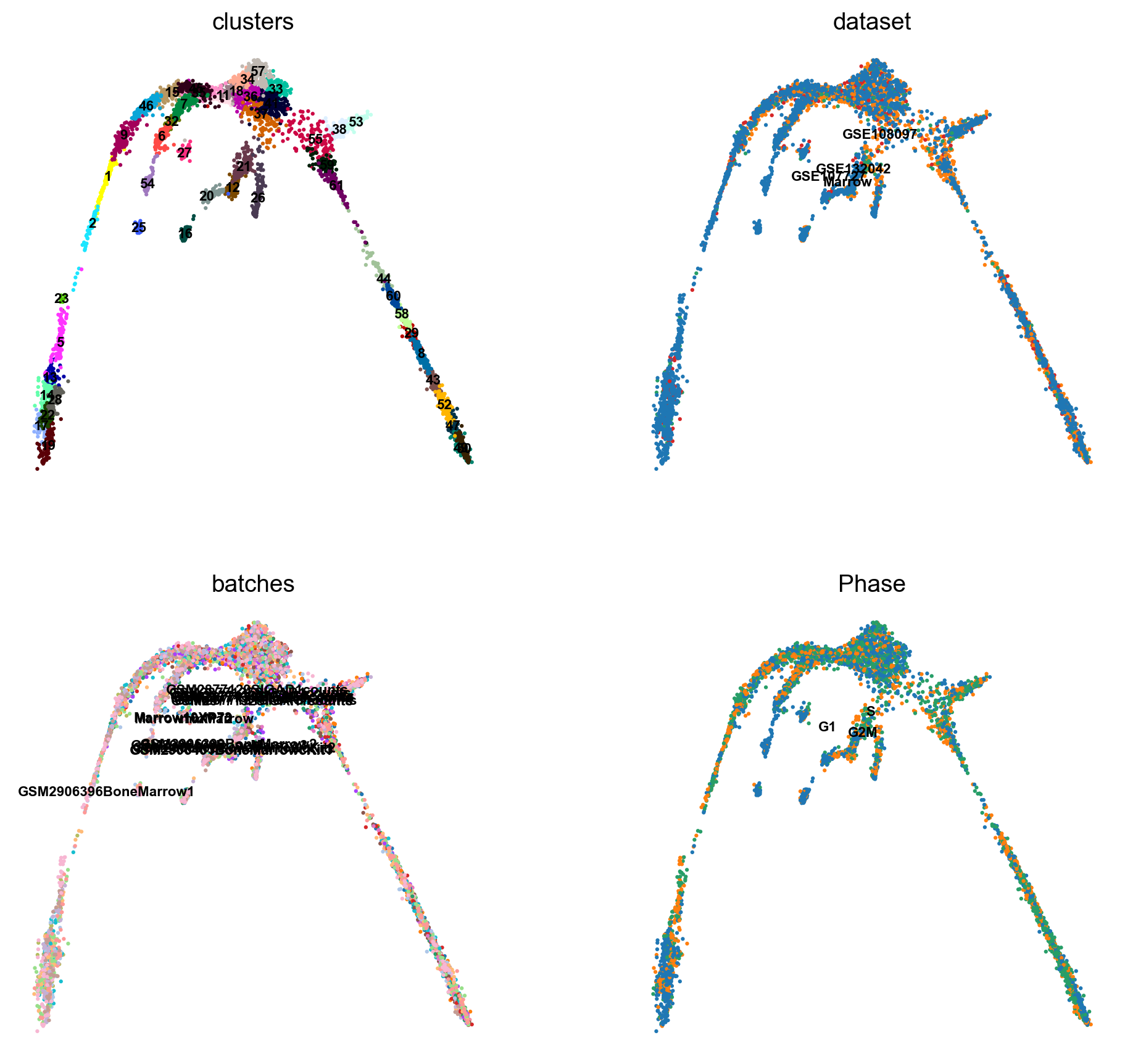
There is a umap and clusters provided with the object, first plot some information from the previous analysis onto the umap.
sc.pl.umap(adata, color = ['clusters','dataset','batches','Phase'],legend_loc = 'on data', legend_fontsize = 'xx-small', ncols = 2)
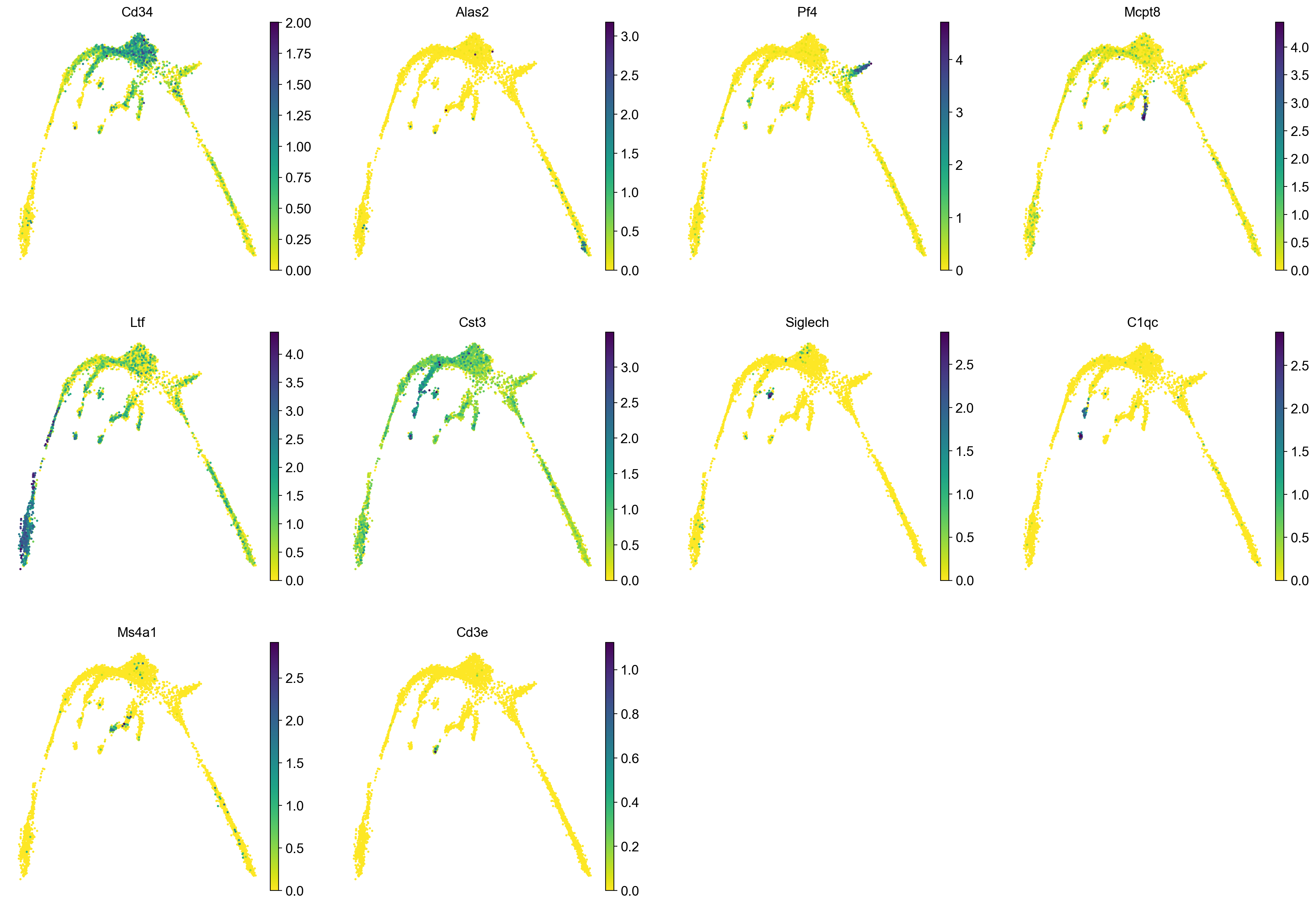
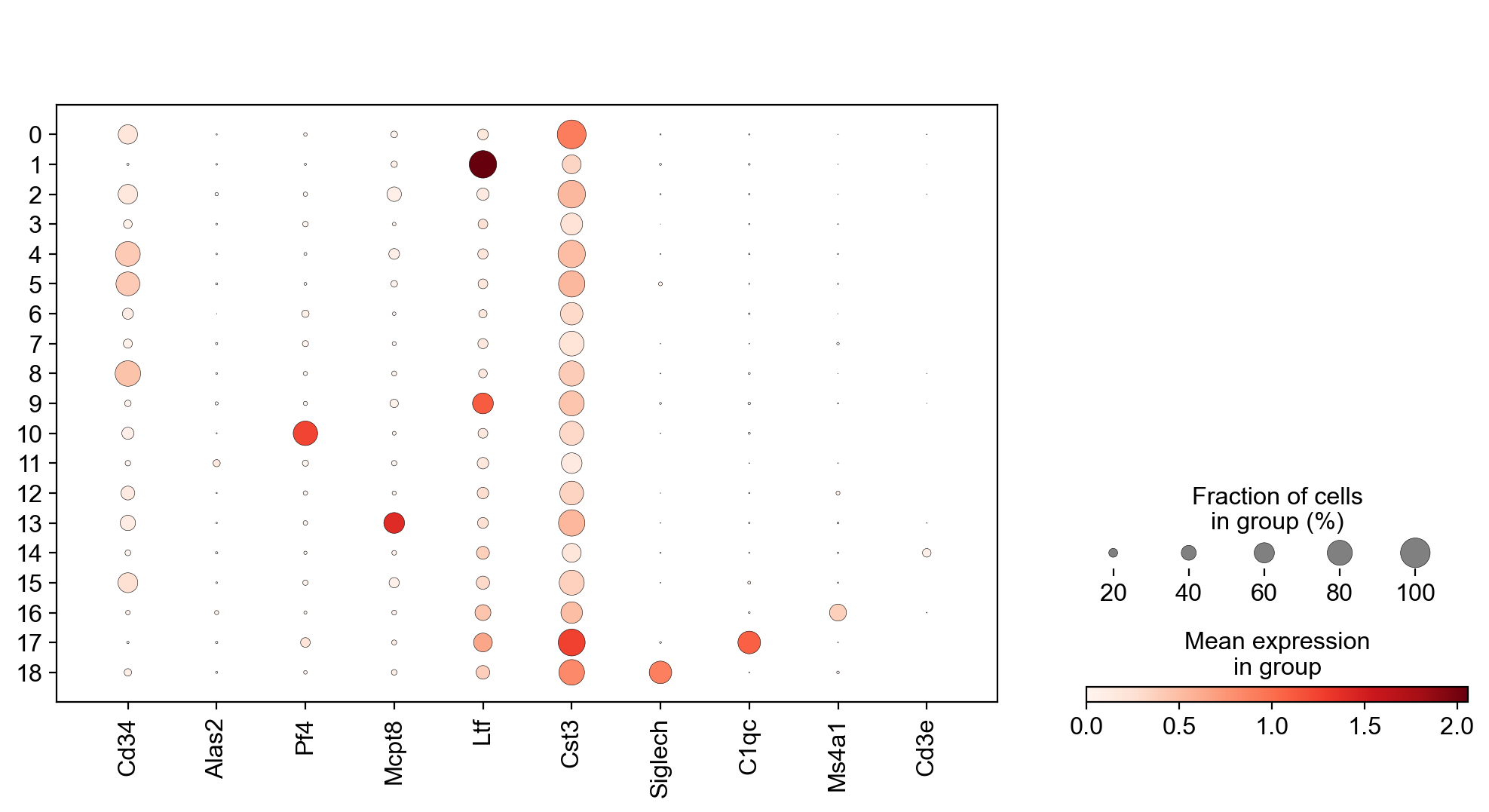
It is crucial that you performing analysis of a dataset understands what is going on, what are the clusters you see in your data and most importantly How are the clusters related to each other?. Well, let’s explore the data a bit. With the help of this table, write down which cluster numbers in your dataset express these key markers.
| Marker | Cell Type |
|---|---|
| Cd34 | HSC progenitor |
| Ms4a1 | B cell lineage |
| Cd3e | T cell lineage |
| Ltf | Granulocyte lineage |
| Cst3 | Monocyte lineage |
| Mcpt8 | Mast Cell lineage |
| Alas2 | RBC lineage |
| Siglech | Dendritic cell lineage |
| C1qc | Macrophage cell lineage |
| Pf4 | Megakaryocyte cell lineage |
markers = ["Cd34","Alas2","Pf4","Mcpt8","Ltf","Cst3", "Siglech", "C1qc", "Ms4a1", "Cd3e", ]
sc.pl.umap(adata, color = markers, use_raw = False, ncols = 4)
Rerun analysis in Scanpy¶
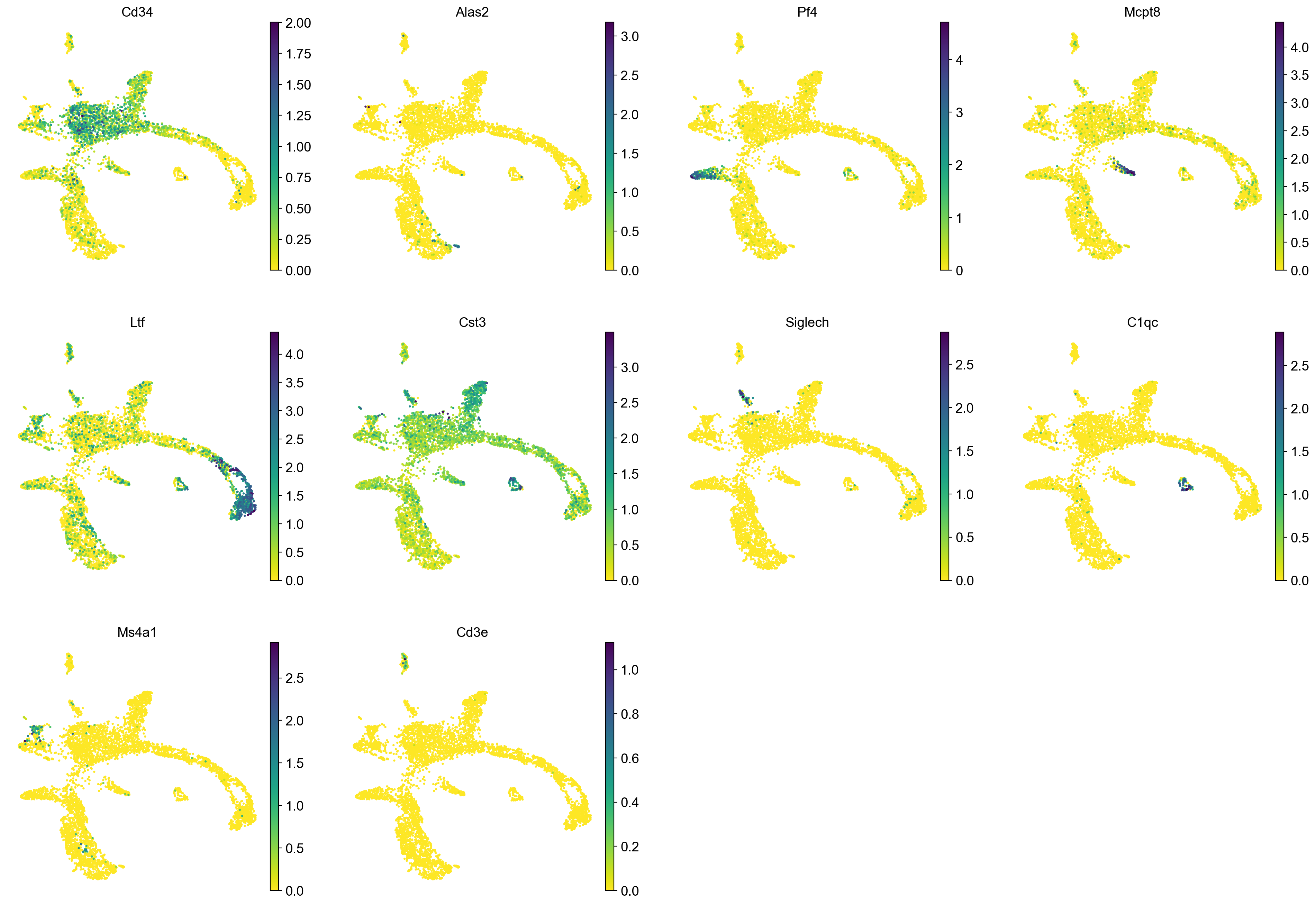
Redo clustering and umap using the basic Scanpy pipeline. Use the provided "X_harmony_Phase" dimensionality reduction as the staring point.
# first, store the old umap with a new name so it is not overwritten
adata.obsm['X_umap_old'] = adata.obsm['X_umap']
sc.pp.neighbors(adata, n_pcs = 30, n_neighbors = 20, use_rep="X_harmony_Phase")
sc.tl.umap(adata, min_dist=0.4, spread=3)
#sc.tl.umap(adata, min_dist=0.6, spread=1.5)
sc.pl.umap(adata, color = ['clusters'],legend_loc = 'on data', legend_fontsize = 'xx-small', edges = True)
sc.pl.umap(adata, color = markers, use_raw = False, ncols = 4)
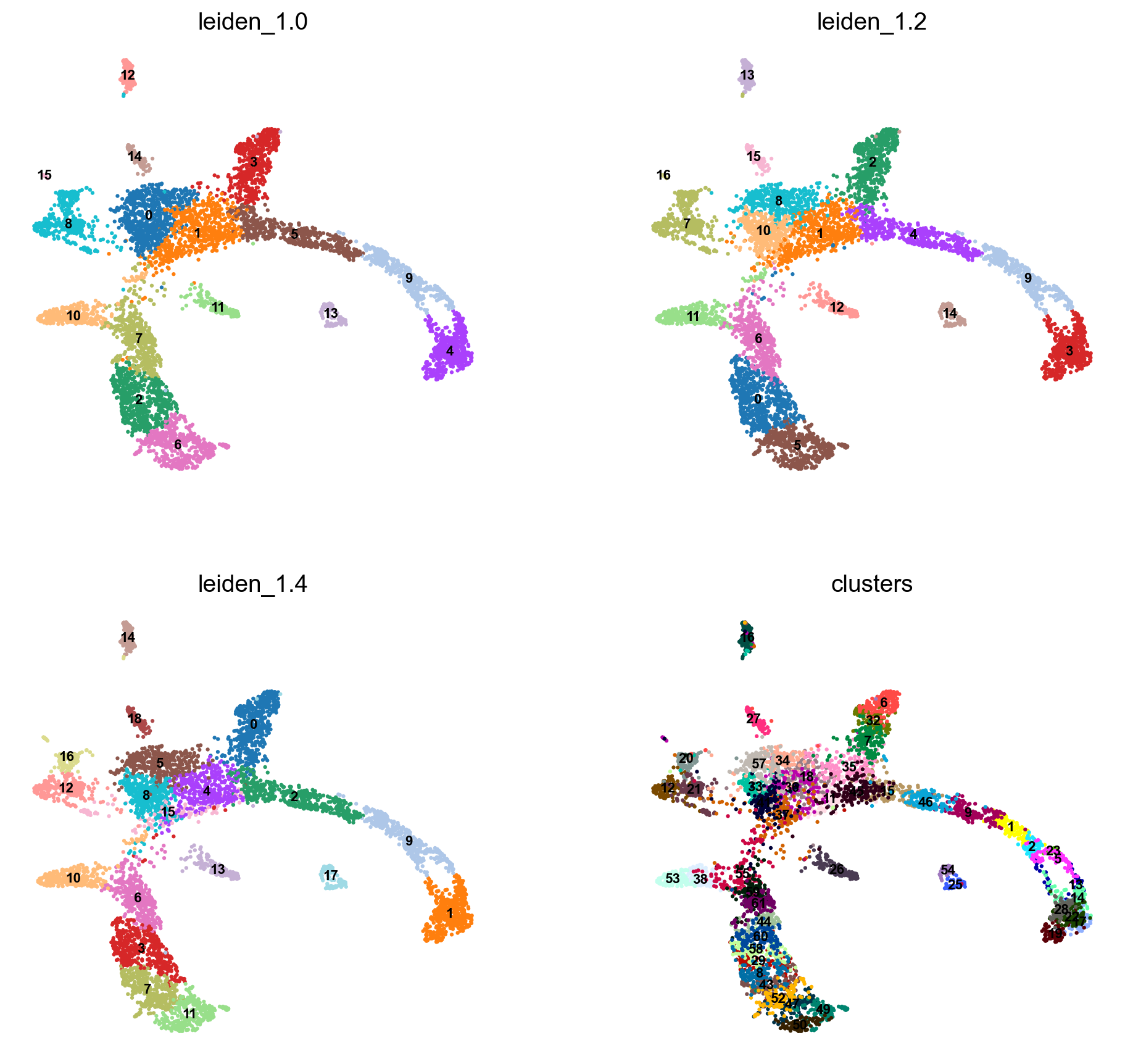
# Redo clustering as well
sc.tl.leiden(adata, key_added = "leiden_1.0", resolution = 1.0) # default resolution in 1.0
sc.tl.leiden(adata, key_added = "leiden_1.2", resolution = 1.2) # default resolution in 1.0
sc.tl.leiden(adata, key_added = "leiden_1.4", resolution = 1.4) # default resolution in 1.0
#sc.tl.louvain(adata, key_added = "leiden_1.0") # default resolution in 1.0
sc.pl.umap(adata, color = ['leiden_1.0', 'leiden_1.2', 'leiden_1.4','clusters'],legend_loc = 'on data', legend_fontsize = 'xx-small', ncols =2)
_, ax = plt.subplots(figsize=(12, 6))
sc.pl.dotplot(adata, var_names=markers, groupby="leiden_1.4", use_raw = False, ax = ax)
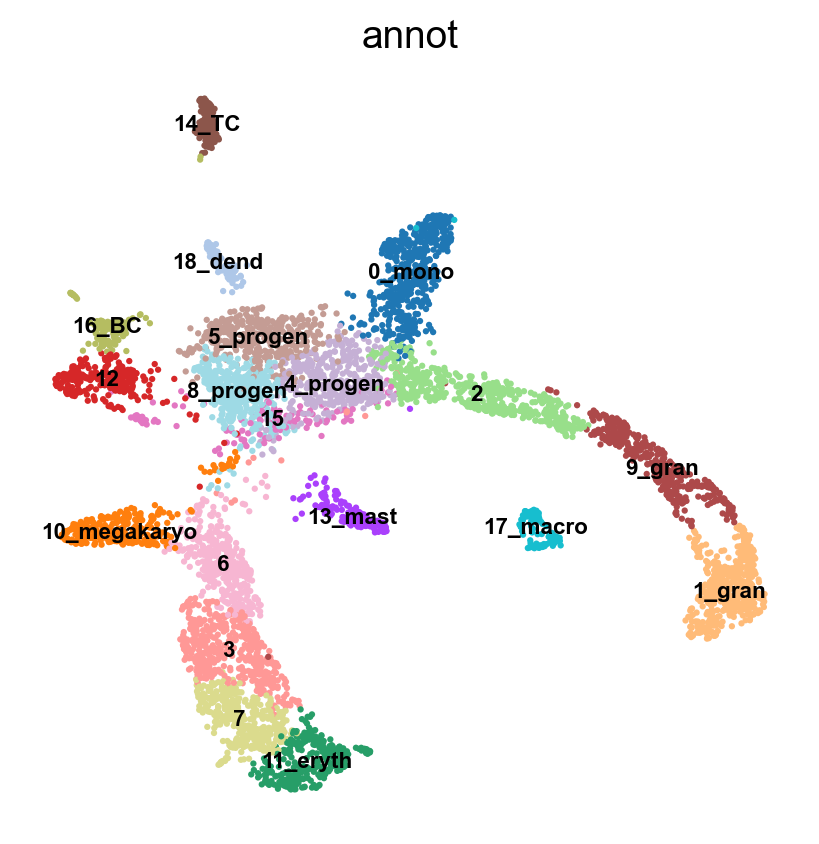
#Rename clusters with really clear markers, the rest are left unlabelled.
annot = pd.DataFrame(adata.obs['leiden_1.4'].astype('string'))
annot[annot['leiden_1.4'] == '10'] = '10_megakaryo' #Pf4
annot[annot['leiden_1.4'] == '17'] = '17_macro' #C1qc
annot[annot['leiden_1.4'] == '11'] = '11_eryth' #Alas2
annot[annot['leiden_1.4'] == '18'] = '18_dend' #Siglech
annot[annot['leiden_1.4'] == '13'] = '13_mast' #Mcpt8
annot[annot['leiden_1.4'] == '0'] = '0_mono' #Cts3
annot[annot['leiden_1.4'] == '1'] = '1_gran' #Ltf
annot[annot['leiden_1.4'] == '9'] = '9_gran'
annot[annot['leiden_1.4'] == '14'] = '14_TC' #Cd3e
annot[annot['leiden_1.4'] == '16'] = '16_BC' #Ms4a1
annot[annot['leiden_1.4'] == '8'] = '8_progen' # Cd34
annot[annot['leiden_1.4'] == '4'] = '4_progen'
annot[annot['leiden_1.4'] == '5'] = '5_progen'
adata.obs['annot']=annot['leiden_1.4'].astype('category')
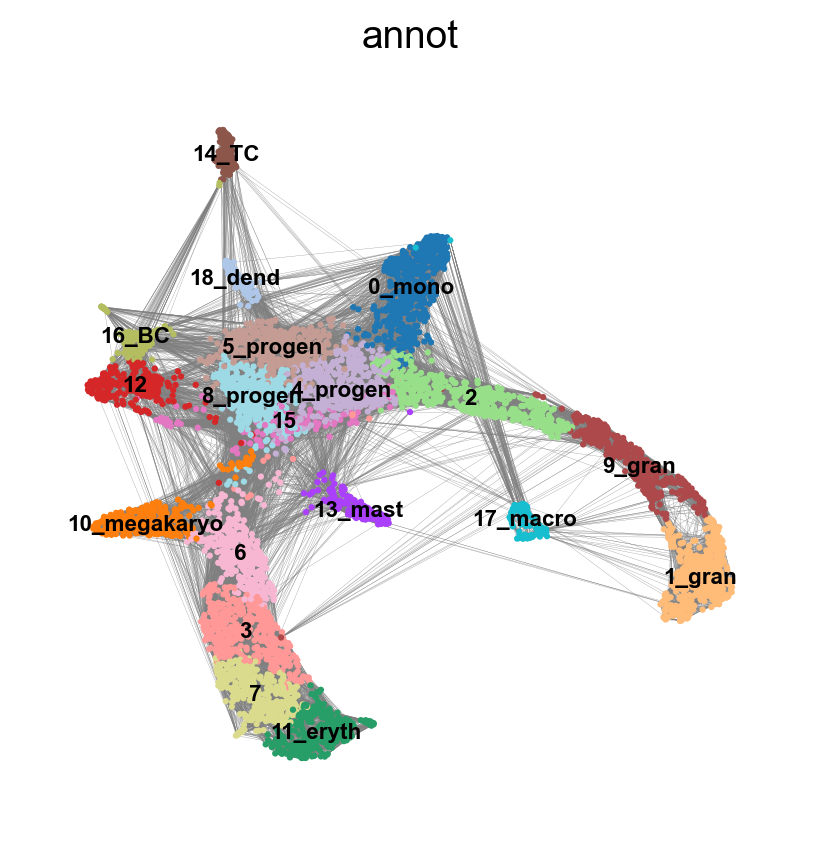
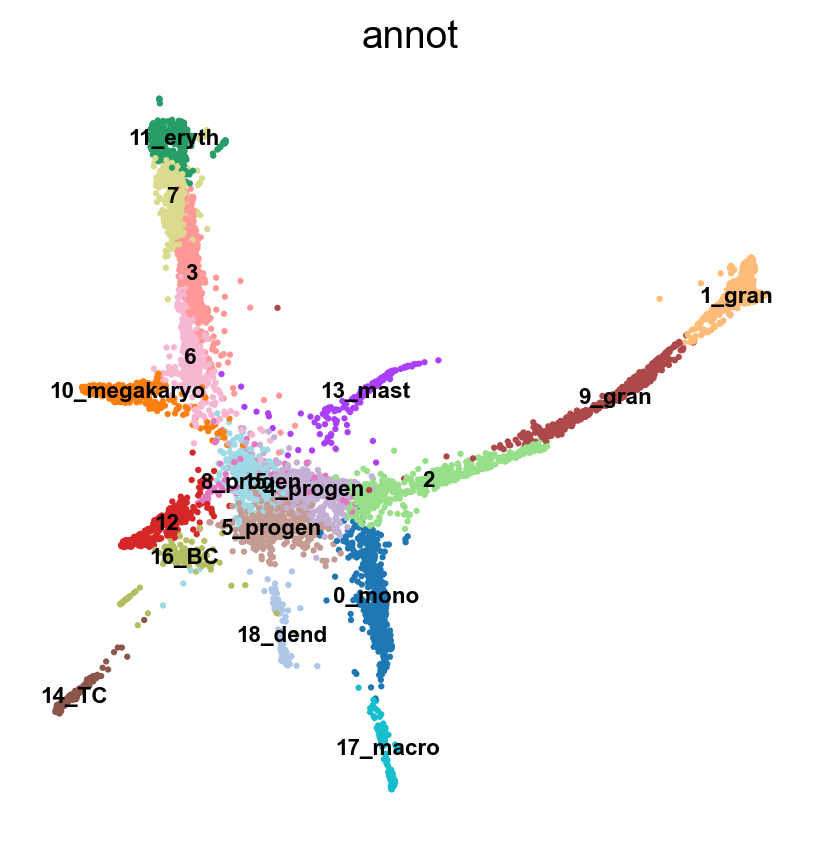
sc.pl.umap(adata, color = 'annot',legend_loc = 'on data', legend_fontsize = 'xx-small', ncols =2)
annot.value_counts()
#type(annot)
# astype('category')
# plot onto the Seurat embedding:
sc.pl.embedding(adata, basis='X_umap_old', color = 'annot',legend_loc = 'on data', legend_fontsize = 'xx-small', ncols =2)
Run PAGA¶
Use the clusters from leiden clustering with leiden_1.4 and run PAGA. First we create the graph and initialize the positions using the umap.
# use the umap to initialize the graph layout.
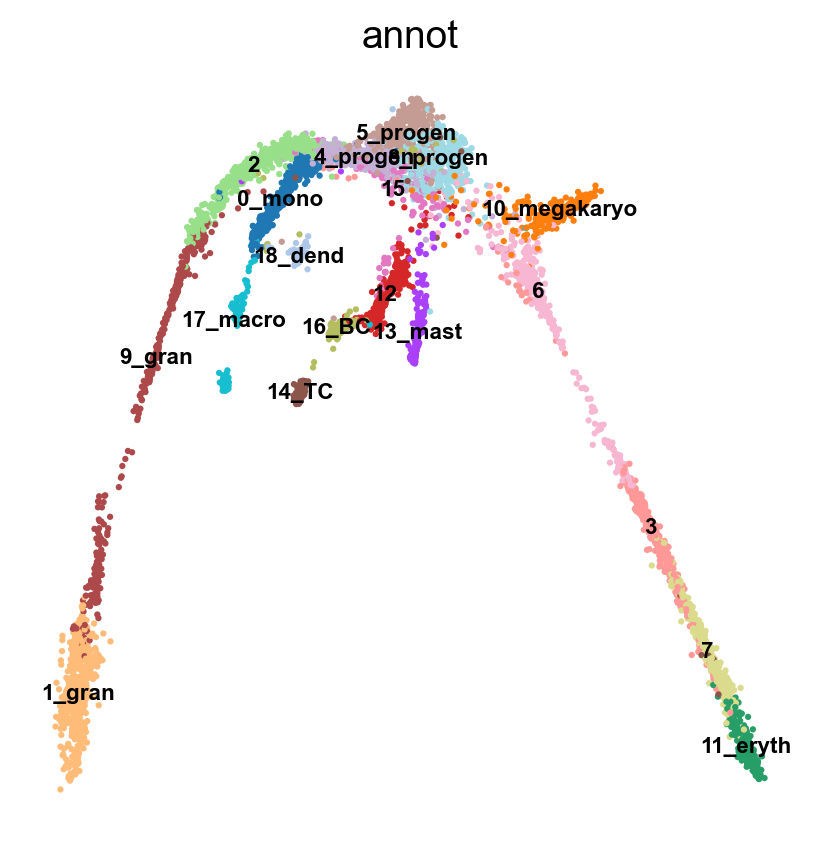
sc.tl.draw_graph(adata, init_pos='X_umap')
sc.pl.draw_graph(adata, color='annot', legend_loc='on data', legend_fontsize = 'xx-small')
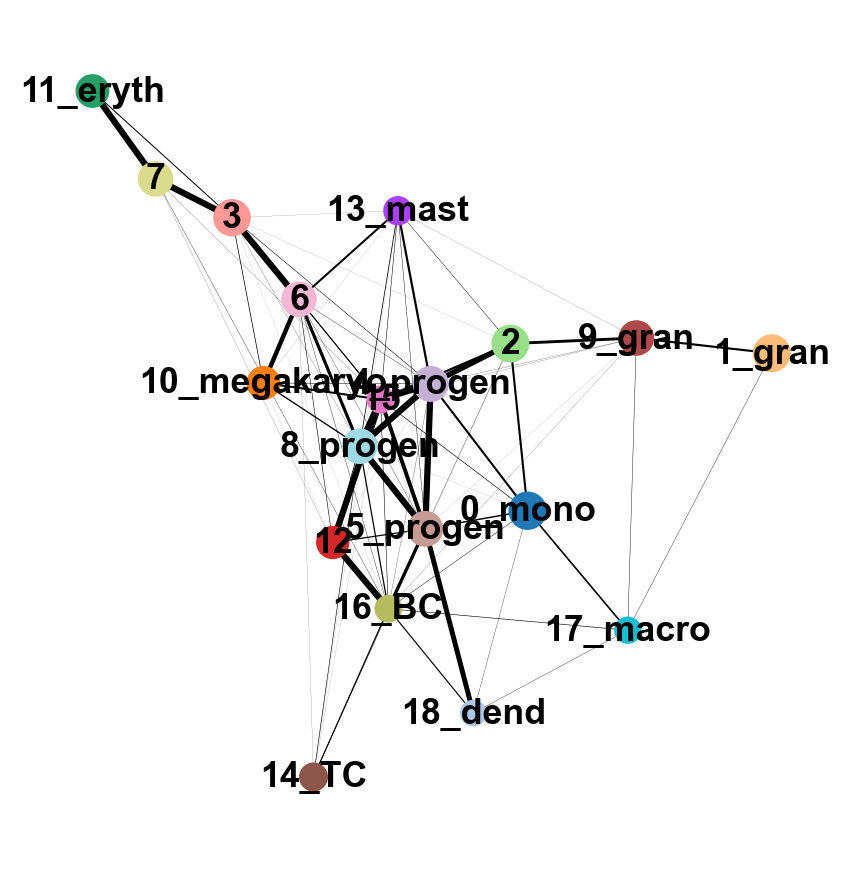
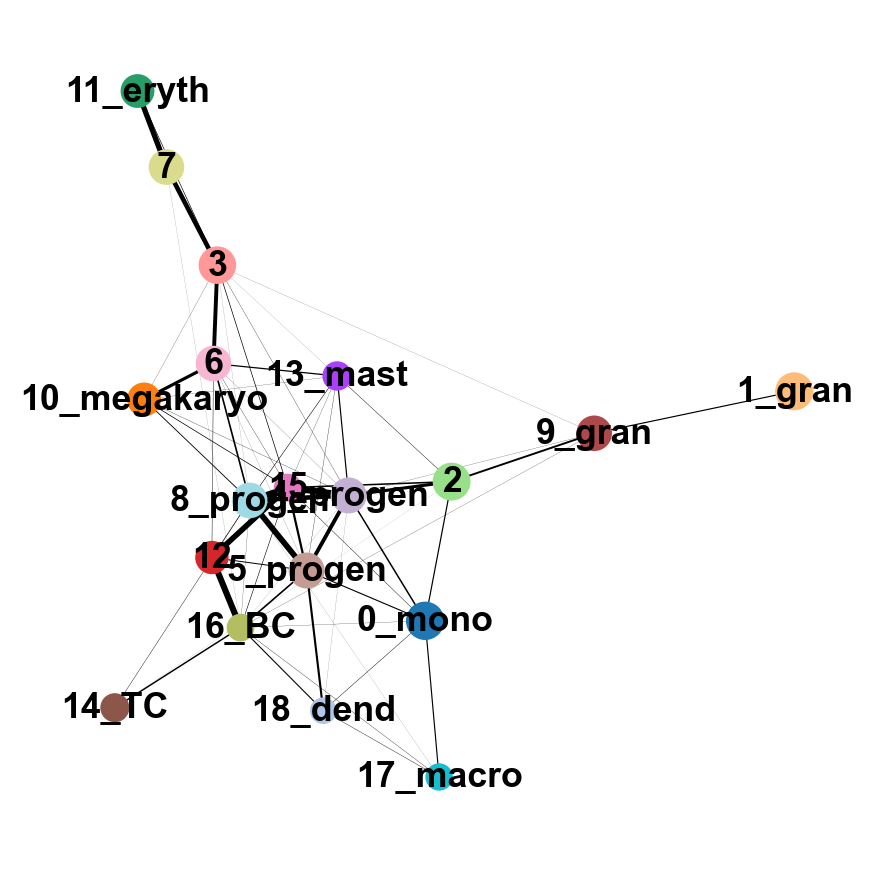
sc.tl.paga(adata, groups='annot')
sc.pl.paga(adata, color='annot', edge_width_scale = 0.3)
As you can see, we have edges between many clusters that we know are are unrelated, so we may need to clean up the data a bit more.
Data pre-processing prior trajectory inference¶
First, lets explore the graph a bit. So we plot the umap with the graph connections on top.
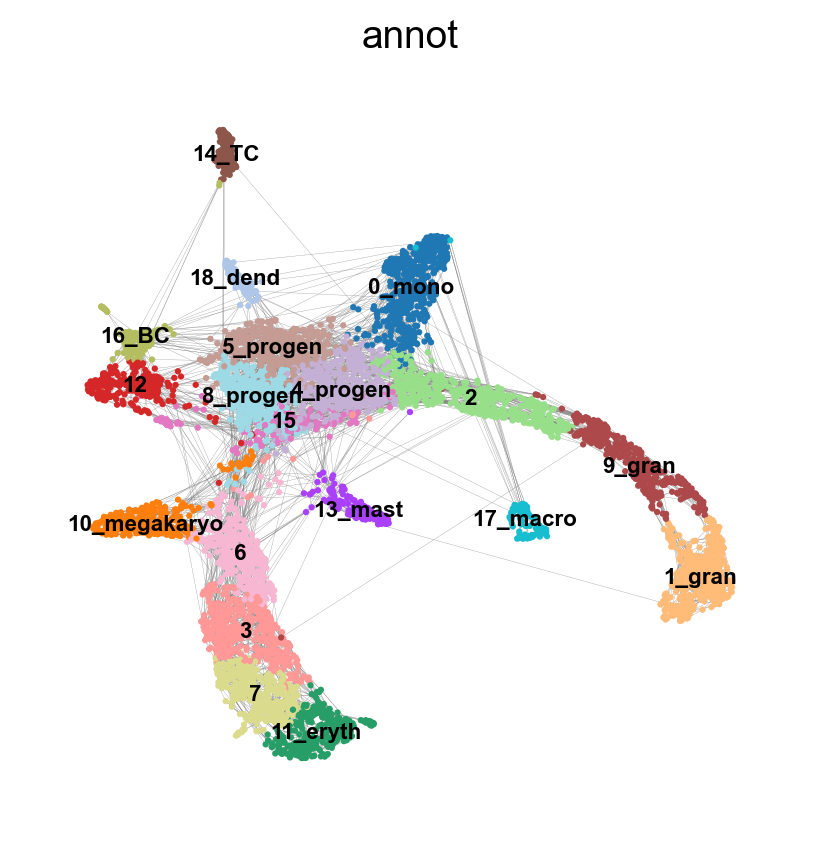
sc.pl.umap(adata, edges=True, color = 'annot', legend_loc= 'on data', legend_fontsize= 'xx-small')
We have many edges in the graph between unrelated clusters, so lets try with fewer neighbors.
sc.pp.neighbors(adata, n_neighbors=5, use_rep = 'X_harmony_Phase', n_pcs = 30)
sc.pl.umap(adata, edges=True, color = 'annot', legend_loc= 'on data', legend_fontsize= 'xx-small')
Rerun PAGA again on the data
sc.tl.draw_graph(adata, init_pos='X_umap')
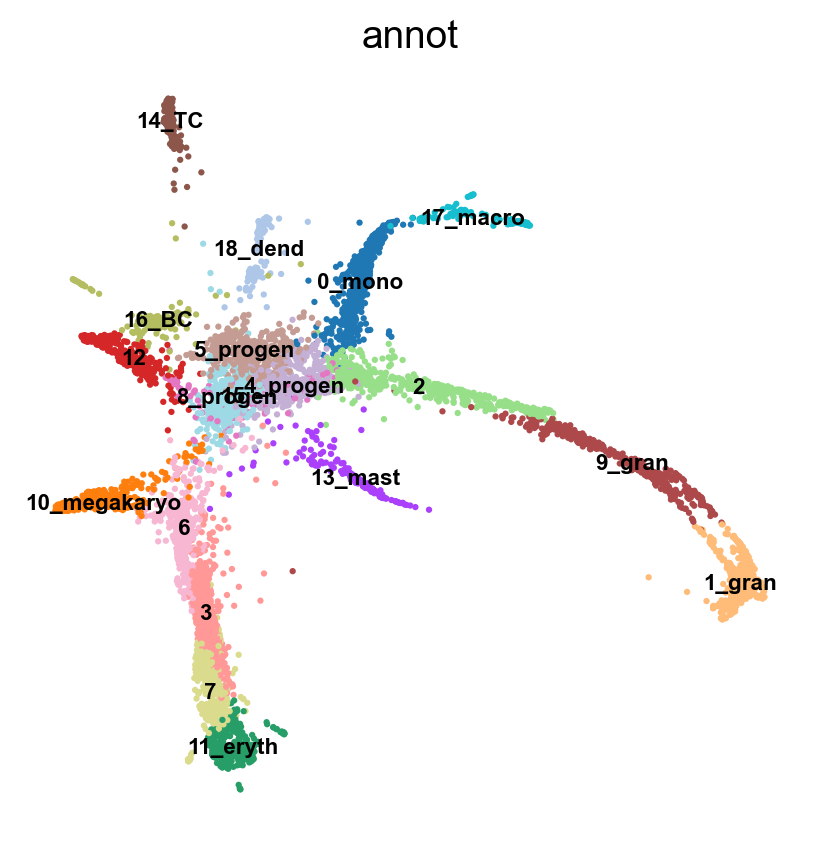
sc.pl.draw_graph(adata, color='annot', legend_loc='on data', legend_fontsize = 'xx-small')
sc.tl.paga(adata, groups='annot')
sc.pl.paga(adata, color='annot', edge_width_scale = 0.3)
Recomputing the embedding using PAGA-initialization¶
The following is just as well possible for a UMAP.
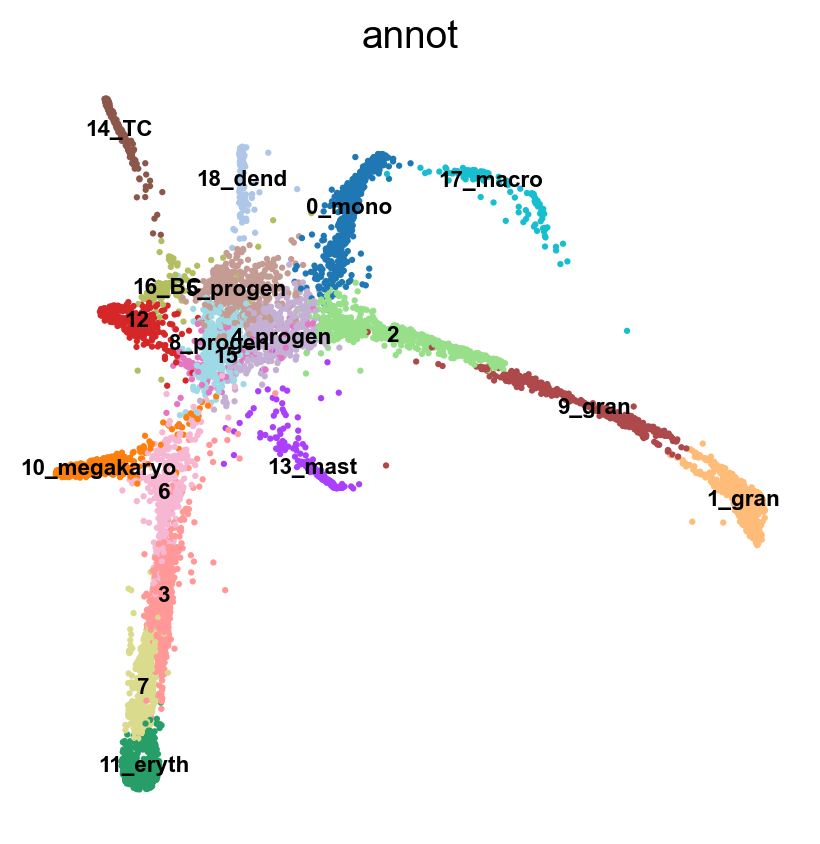
sc.tl.draw_graph(adata, init_pos='paga')
Now we can see all marker genes also at single-cell resolution in a meaningful layout.
sc.pl.draw_graph(adata, color=['annot'], legend_loc='on data', legend_fontsize= 'xx-small')
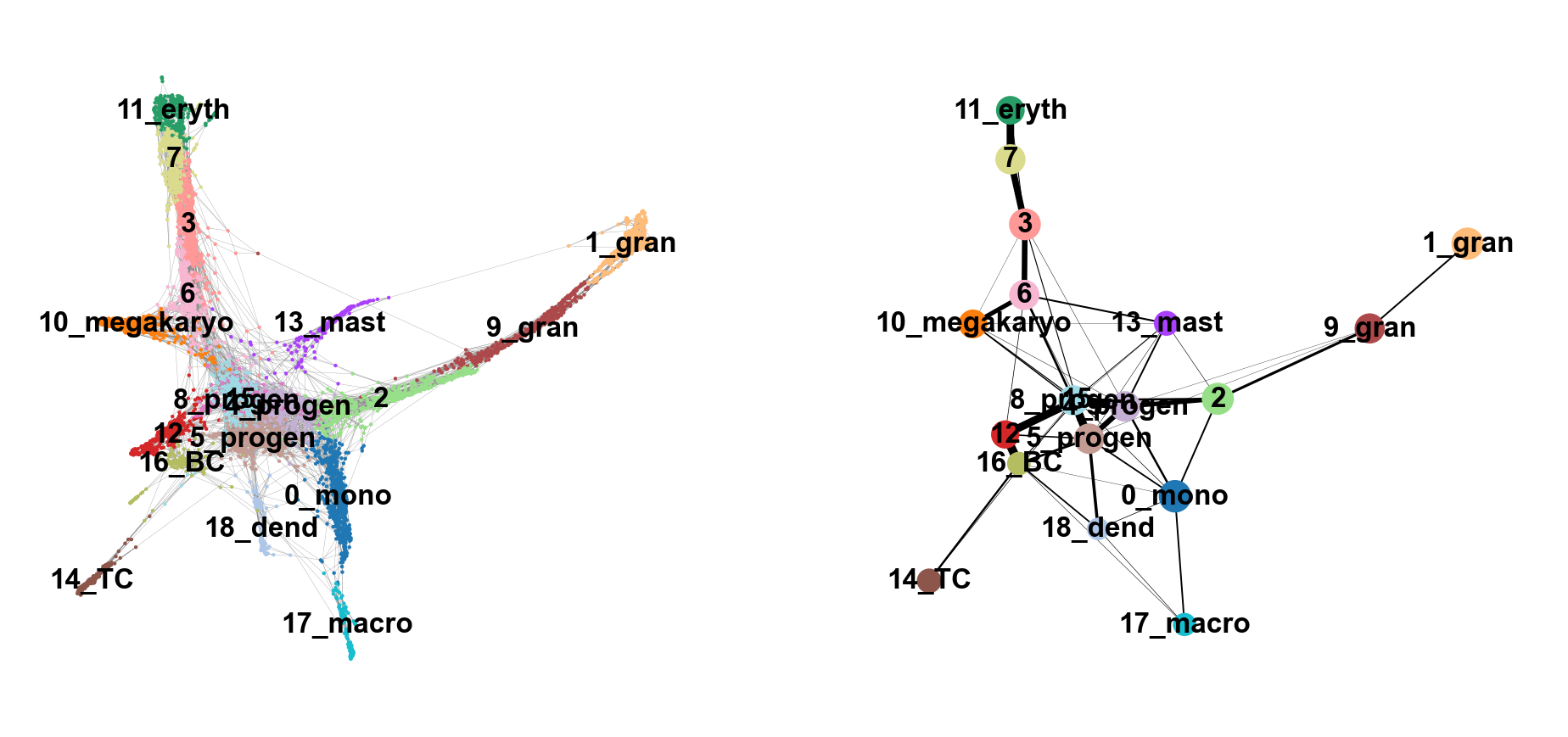
Compare the 2 graphs
sc.pl.paga_compare(
adata, threshold=0.03, title='', right_margin=0.2, size=10, edge_width_scale=0.5,
legend_fontsize=12, fontsize=12, frameon=False, edges=True, save=True)
Reconstructing gene changes along PAGA paths for a given set of genes¶
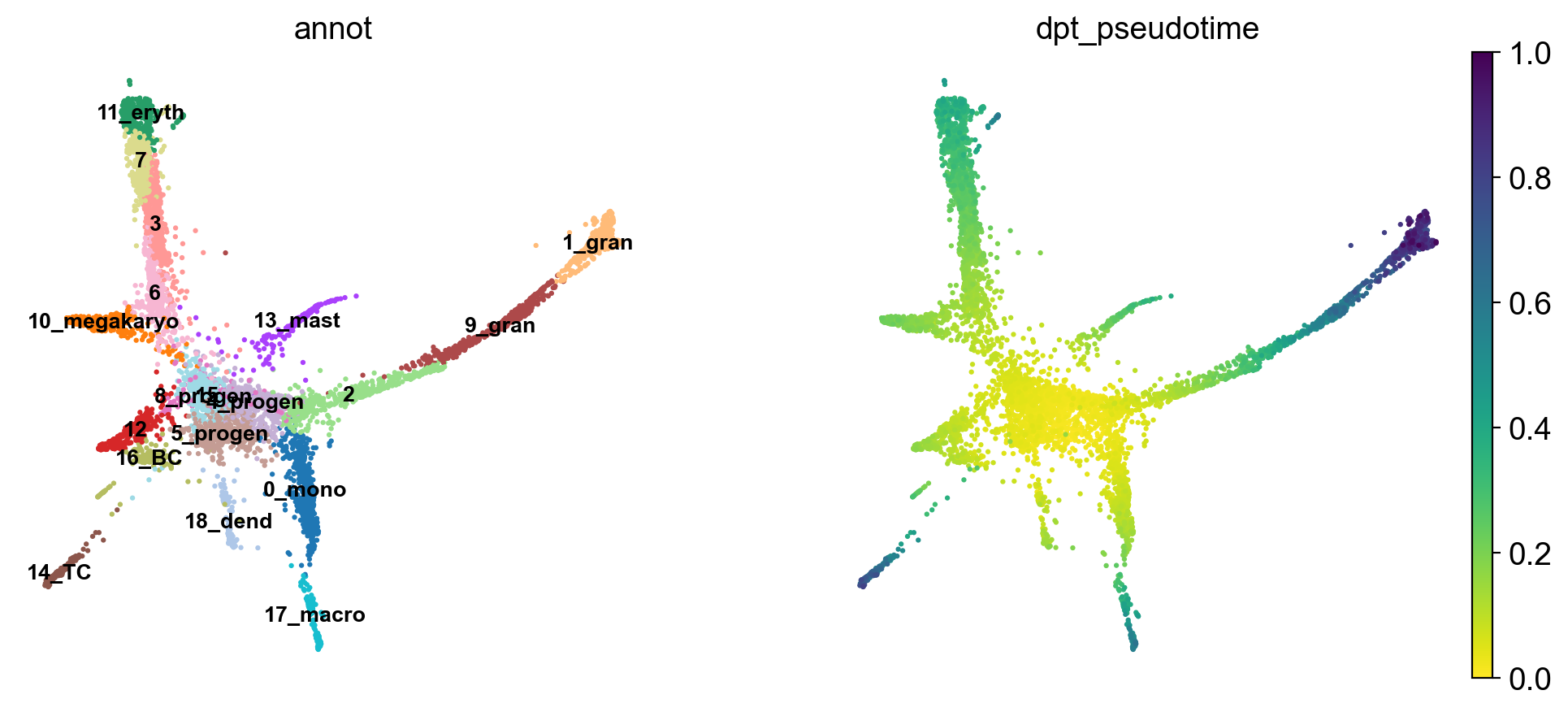
Choose a root cell for diffusion pseudotime. We have 3 progenitor clusters, but cluster 5 seems the most clear.
adata.uns['iroot'] = np.flatnonzero(adata.obs['annot'] == '5_progen')[0]
sc.tl.dpt(adata)
Use the full raw data for visualization.
sc.pl.draw_graph(adata, color=['annot', 'dpt_pseudotime'], legend_loc='on data', legend_fontsize= 'x-small')
By looking at the different know lineages and the layout of the graph we define manually some paths to the graph that corresponds to spcific lineages.
# Define paths
paths = [('erythrocytes', ['5_progen', '8_progen', '6','3','7', '11_eryth']),
('lympoid', ['5_progen', '12', '16_BC', '14_TC']),
('granulo', ['5_progen', '4_progen', '2', '9_gran', '1_gran']),
('mono', ['5_progen','4_progen','0_mono','18_dend','17_macro'])
]
adata.obs['distance'] = adata.obs['dpt_pseudotime']
!mkdir write
Then we select some genes that can vary in the lineages and plot onto the paths.
gene_names = ['Gata2', 'Gata1', 'Klf1', 'Epor', 'Hba-a2', # erythroid
'Elane', 'Cebpe', 'Gfi1', # neutrophil
'Irf8', 'Csf1r', 'Ctsg', # monocyte
'Itga2b','Prss34','Cma1','Procr', # Megakaryo,Basophil,Mast,HPC
'C1qc','Siglech','Ms4a1','Cd3e','Cd34']
_, axs = pl.subplots(ncols=4, figsize=(10, 4), gridspec_kw={'wspace': 0.05, 'left': 0.12})
pl.subplots_adjust(left=0.05, right=0.98, top=0.82, bottom=0.2)
for ipath, (descr, path) in enumerate(paths):
_, data = sc.pl.paga_path(
adata, path, gene_names,
show_node_names=False,
ax=axs[ipath],
ytick_fontsize=12,
left_margin=0.15,
n_avg=50,
annotations=['distance'],
show_yticks=True if ipath==0 else False,
show_colorbar=False,
color_map='Greys',
groups_key='annot',
color_maps_annotations={'distance': 'viridis'},
title='{} path'.format(descr),
return_data=True,
use_raw = False,
show=False)
data.to_csv('./write/paga_path_{}.csv'.format(descr))
pl.savefig('./figures/paga_path.pdf')
pl.show()
Your turn¶
As you can see, we can manipulate the trajectory quite a bit by selecting different number of neighbors, components etc. to fit with our assumptions on the development of these celltypes.
Please explore furhter how you can tweak the trajectory. For instance, can you create a PAGA trajectory using the orignial umap from Seurat instead? Hint, you first need to compute the neighbors on the umap