class: center, middle, inverse, title-slide .title[ # Clustering ] .subtitle[ ## Workshop on RNA-Seq ] .author[ ### <b>Nima Rafati</b> ] .institute[ ### NBIS, SciLifeLab ] --- --- name: intro ## Clustering - What is clustering? - Clustering is an approach to classify/group data points. - Why do we use clustering? - For exploring the data - To discover patterns in our data set - Identify outliers --- name: clustering-method ## Clustering Methods - Centroid-based - Density-based - Distribution-based - Hierarchical-based ## Steps: In short all clustering approach follows these steps: - Calculate distance between data points - Group \| cluster the data based on similarities --- name: distance-metrics ## Distance can be measured in: - In multidimensional space (raw data) - In reduced space (i.e. top PCs) --- name: Euclidean ## Euclidean distance - Euclidean distance is a straight line between two points $$ c^2 = a^2 + b^2 $$ <img src="slide_clustering_files/figure-html/euclidean-distance-1.png" style="display: block; margin: auto;" /> --- name: Manhattan-distance ## Manhattan distance - Manhattan distance $$ a + b $$ <img src="slide_clustering_files/figure-html/manhattan-distance-1.png" style="display: block; margin: auto;" /> --- name: Inverted-pairwise-correlations ## Inverted pairwise correlations - Inverted pairwise correlations $$ dist = -(cor -1) / 2 $$ <img src="slide_clustering_files/figure-html/inverted-pairwise-correlation-1.png" style="display: block; margin: auto;" /> --- name: Mahalanobis-Distance ## Mahalanobis Distance - Despite of previous approach which was based on distance between data points, this method measures the distance between a data point and a distribution. <img src="data/Mahalanobis.png" width="60%" style="display: block; margin: auto;" /> --- name: Centroid-based1 ## Centroid-based: K-means clustering - One of the most commonly used clustering methods - In this method the distance between data points and centroids is calculated - Each data point is assigned to a cluster based on Euclidean distance from centroid. - Dependent on number of K (clusters) new centroids are created <!-- <div style="text-align: center;"> --> <!-- <img src="data/kmeans_3.gif" alt="Alt text for the GIF" style="width: 65%; height: auto;"> --> <!-- </div> --> --- name: Centroid-based2 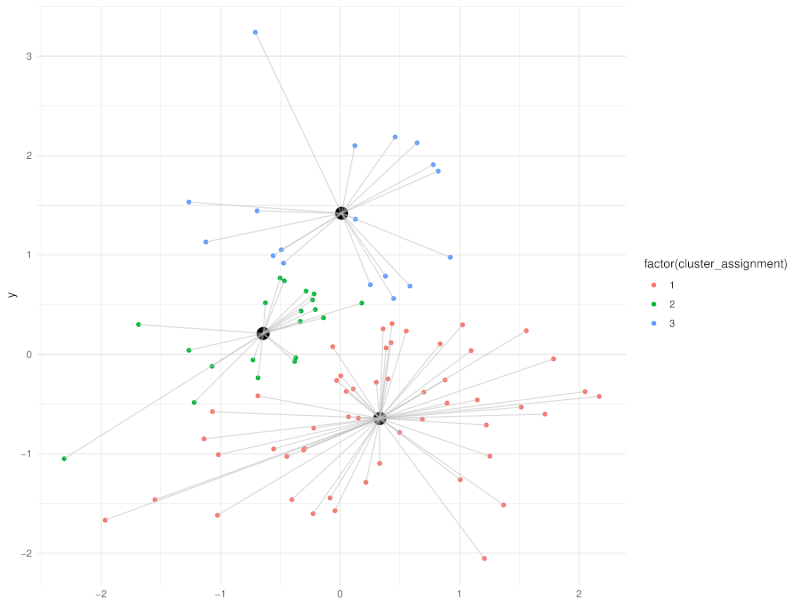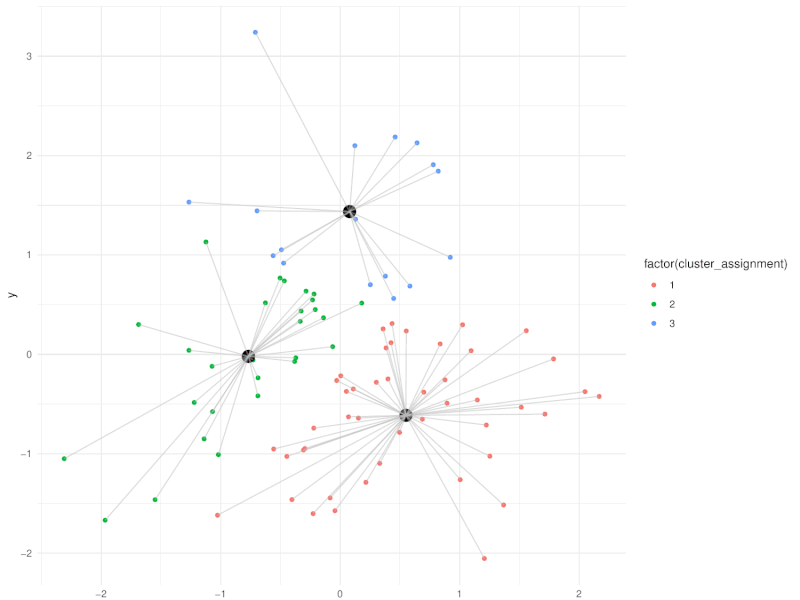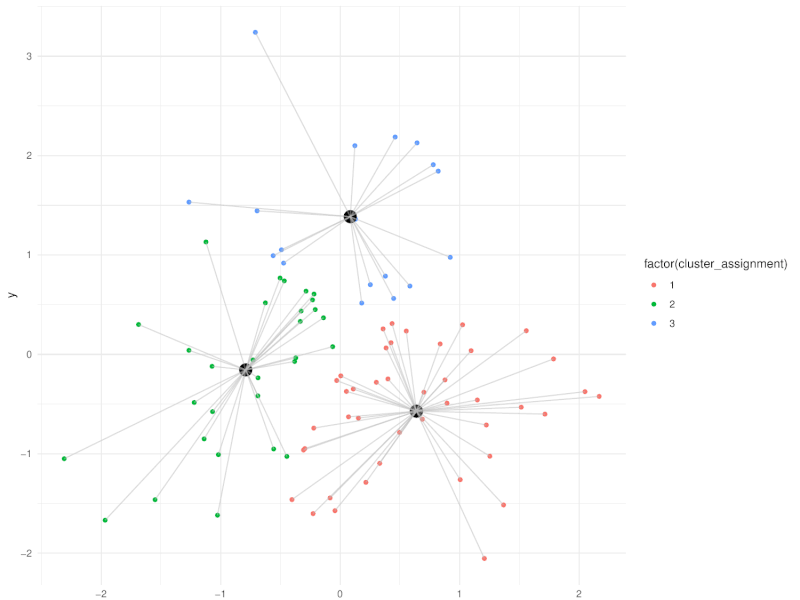{width=50%} --- name: Centroid-based3 ## Centroid-based: K-means clustering - One of the most commonly used clustering methods - In this method the distance between data points and centroids is calculated - Each data point is assigned to a cluster based on Euclidean distance from centroid. - Dependent on number of K (clusters) new centroids are created --- name: Centroid-based4 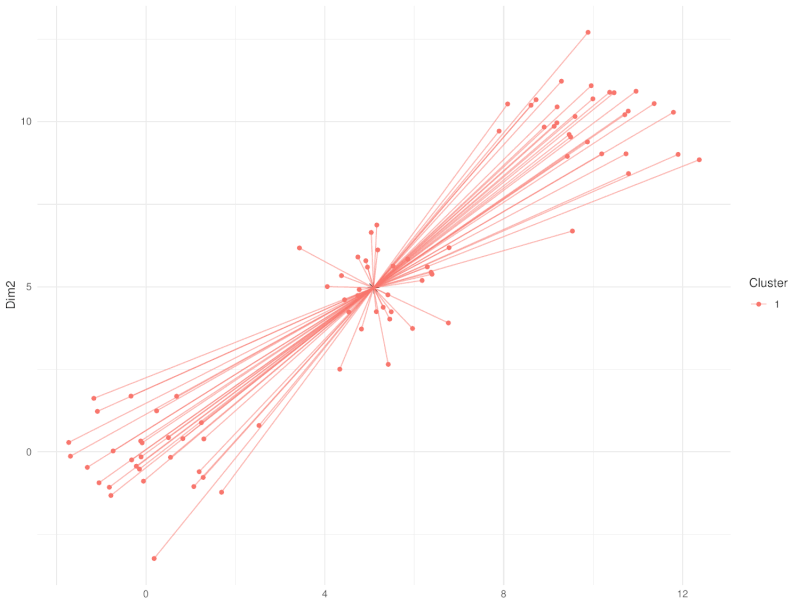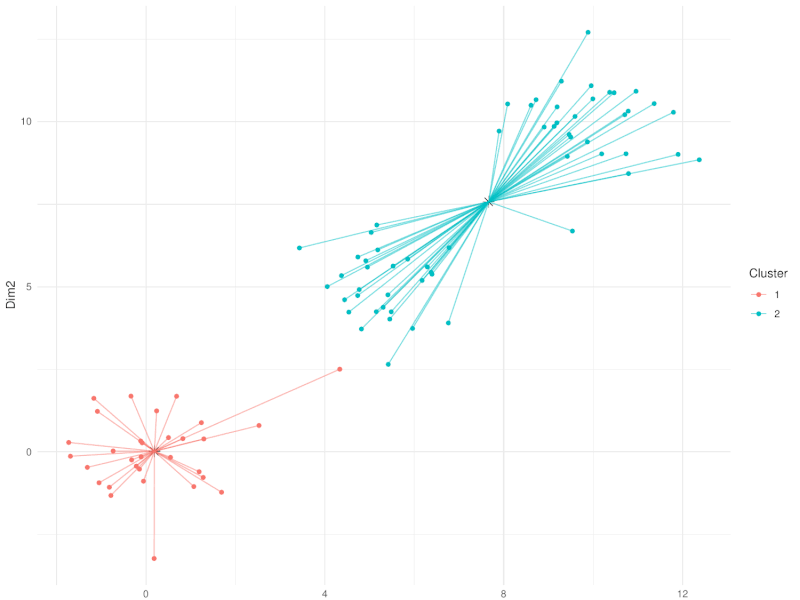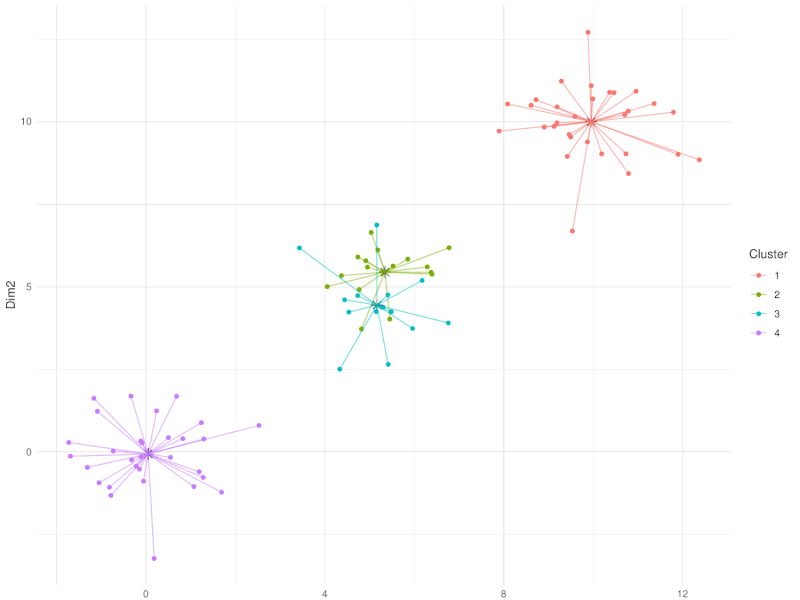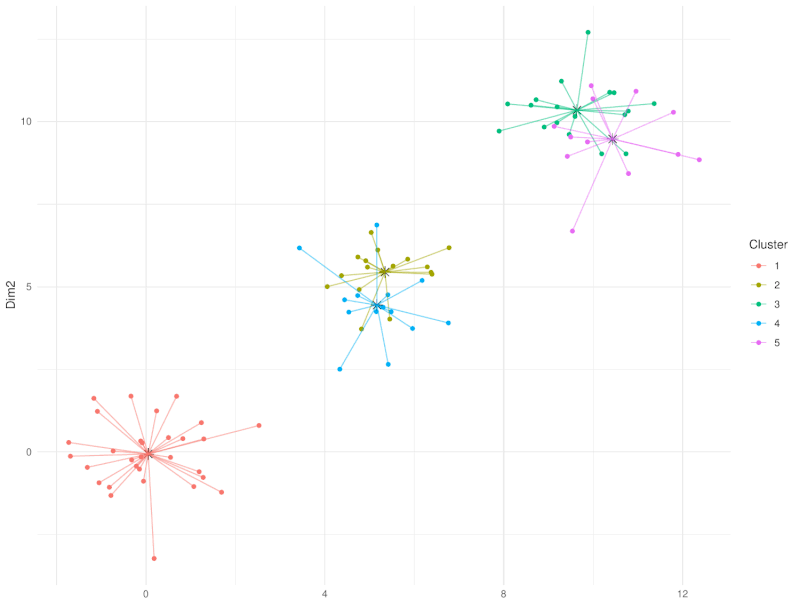{width=50%} --- name: optimal k ## What is optimal K? - The user needs to define the number of clusters: - **Elbow method**. - Gap statistics. - Average Silhouette method <img src="slide_clustering_files/figure-html/optimal-k-1.png" style="display: block; margin: auto;" /> ??? Within-cluster sum of squares (WCSS) is a metric used to quantify the compactness of clusters by measuring the squared distances from each point to its cluster centroid. It serves as a key indicator for determining the optimal number of clusters in a dataset. In the Elbow method, the objective is to identify a suitable number of clusters (k) by locating the point where increases in k result in diminishing reductions in WCSS. This 'elbow' point is considered optimal because beyond it, additional clusters do not significantly enhance the model's performance in terms of intra-cluster compactness. --- name: DBSCAN ## Density-based clustering: DBSCAN - This method identifies regions within your data distribution that exhibits high density of data points. --- name: GMM ## Distribution-based clustering: Guassian Mixture Model (GMM) - It involves modeling the data points with probability distribution. - In this method prior knowledge on distribution of your data is required. If you do not know the distribution of your data try another approach. - You need to specify number of clusters. ??? Distribution-based clustering involves modeling the data points using probability distributions, with the Gaussian Mixture Model (GMM) being one of the most commonly used methods in this category. In a GMM, each cluster is modeled as a Gaussian distribution, and the algorithm iteratively updates the parameters of these distributions (mean, covariance) and the probability of each point belonging to each cluster. --- name: hclust ## Hierarchical-based clustering - This approach creates a tree of clusters. - Well suited for hierarchical data (e.g. taxonomies). - Final output is a dendrogram representing the order decisions at each merge/division of clusters. - Two approaches: - Agglomerative (Bottom-up): All data points are treated as clusters and then joins similar ones. - Divisive (Top-down): All data points are in one large clusters and recursively splits the most heterogeneous clusters. - Number of clusters are decided after generating the tree. --- name: hclust-agglomorative ## Hierarchical-based clustering - Agglomerative clustering <img src="data/HC-agglomerative.png" width="60%" /> --- name: hclust-divisive ## Hierarchical-based clustering - Divisive clustering <img src="data/HC-divisive.png" width="60%" /> --- name: linkage ## Linkage methods. To combine clusters, it's essential to establish their positions relative to one another. The technique used to determine these positions is known as **Linkage**. <img src="data/Linkages.png" width="60%" style="display: block; margin: auto;" /> --- name: linear-clustering-summary ## Summary - For bulk RNASeq you can perform clustering on raw, Z-Score scaled data or on top PC coordinates. - For the sample large size (>10,000) you can perform clustering on PC. For instance in scRNASeq data. - You always need to tune some parameters. - K-means performs poorly on unbalanced data. - In hierarchical clustering, some distance metrics need to be used with a certain linkage method. - Checking clustering Robustness (a.k.a Ensemble perturbations): - Most clustering techniques will cluster random noise. - One way of testing this is by clustering on parts of the data (clustering bootstrapping) - Read more in [Ronan et al (2016) Science Signaling](https://www.science.org/doi/10.1126/scisignal.aad1932?url_ver=Z39.88-2003&rfr_id=ori:rid:crossref.org&rfr_dat=cr_pub%20%200pubmed). --- name: Know more ## Do you want to know more? Please check the following links: - [Avoiding common pitfalls when clustering biological data](https://www.science.org/doi/10.1126/scisignal.aad1932?url_ver=Z39.88-2003&rfr_id=ori:rid:crossref.org&rfr_dat=cr_pub%20%200pubmed) - [Clustering with Scikit with GIFs](https://dashee87.github.io/data%20science/general/Clustering-with-Scikit-with-GIFs/) (Note, this is based on python but provide nice illustration). --- name: end_slide class: end-slide, middle count: false # Thank you. Questions?