Overview
Teaching: 15 min
Exercises: 0 minQuestions
What are research data repositories?
Why submit my data to a repository?
How do I find a suitable repository?
Objectives
Explain why data should be publicly available.
Explain different types of repositories and how to find a suitable one.
What are research data repositories?
Traditionally ‘all’ research outputs were put in the published article only, resulting in that the findings often where difficult to reproduce let alone reuse, or put in a local web server with limited longevity. Fortunately times are changing and nowadays there is a pletora of repositories to choose between, in order to make research as easy and widely available as possible.
Why submit your datasets to a repository?
Why should you care? Submitting your data to a repository likely takes a lot of time, time you could spend on doing research, and employers as well as funders are only interested in how many articles you have published, not how many datasets you have published. Well, times are indeed changing, there are many institutions with data policies on sharing, and funders want maximum value for their invested money. Besides, if you do not share your data, you cannot ask of others to share either, it’s as simple as that. You could argue that you do share your data if someone asks for it (aka the (in)famous phrase ‘available upon request’) but repositories provides the technical solution to FAIR data:
- Findable by being assigned a persistent identifier, and by being described with rich metadata.
- Accessible by being put in a resource that is searchable, and enables easy access via internet
- Interoperable by using standard format and language to represent both the data and its metadata
- Reusable by fulfilling the F, A, and I, and by having a clear and accessible data usage license
Hence, by submitting data to a repository, your data becomes FAIR and you do not have to provide a solution on your own.
Reasons for sharing in a repository:
- Open Science & FAIR - To meet the requirements from funders and society on Open Science & FAIR
- Reproducibility - So that your published research results can be reproduced
- Trail of evidence - To provide a provenance of the data
- Reusability / 3rd party access - To give others easy access to your data
- Archival purposes - Research data should be available for as long as it is useful to someone
- Publication of paper requires it - Nowadays most publishers require you to submit the data to a repository when publishing a paper
What research outputs should be submitted?
Apart from the data itself, everything necessary to understand, reproduce and reuse the data should be submitted to a repository:
- Raw data: this is the data that comes straight from the instrument, e.g. RNA sequences in fastq format
- Processed & analysed data: this is the data where some type of analysis or processing has been done, e.g. normalization, removal of outliers, expression measurements, statistics, annotation
- Metadata: this is the description of the raw and processed data, e.g. in the form of minimum information to reproduce the data, sample information, precise protocols, analysis scripts and code, etc
How do I find a suitable repository?
As mentioned, there is now a pletora of repositories, with varying reach (impact) and depth (rich metadata descriptions), and they are often divided into three categories: domain-specific, general purpose, and in-house/institutional.
Domain-specific repositories
This type of repository focuses on specific data types and is typically the best choice if you can find one that is suitable for your research data. It will reach your research community, so that others working in your field can find and reuse your data, and incorporates metadata standards in order to make the data as widely useful as possible. The repositories usually have long-term sustainability plan, i.e. they will be available for a long time, and are typically free of charge.
Repositories for sensitive human data
Sensitive personal data is a special category of personal data as defined by the General Data Protection Regulation (GDPR). Examples of this data type within the biomedical research field are genetic data and health data (sensitive human data). Sensitive human data cannot be made publicly available in ordinary open-access domain specific repositories but can instead be deposited in special repositories such as the European Genome-phenome Archive (EGA) or the Swedish Federated European Genome-phenome Archive (FEGA Sweden). In this case the metadata records are openly accessible but the sensitive data is made available under restricted access. Restricted access means that researchers can request access to the sensitive data through a formalized application procedure.
About sensitive human data
What data is regarded as personal data? Well, that depends on the context. If you only have a name, it is not considered to be personal, and even if you have a name and the city where this person lives, it is likely not personal data. However, if you also have date of birth, this is likely considered as personal data.
What data is regarded as sensitive personal data? Sensitive personal data, as defined by the General Data Protection Regulation (GDPR) (Art. 9), refers to information that is particularly private or sensitive in nature. When it comes to life science research, Health data and Genetic data are the most relevant categories of sensitive personal data. This means that if you are working with DNA sequences, the sequences themselves are sensitive personal data, irrespective of what information you have regarding who’s gene sequence it is.
Another example of sensitive personal data is if we have the information name, city, and date of birth, i.e. typically personal but not sensitive data, and we add disease diagnosis information, this is classified as sensitive personal data.
SciLifeLab RDM guidelines have more guidance when it comes to Research involving human data.
Examples:
- European Nucleotide Archive (ENA) - for genomic sequence data (non-human)
- European Genome Phenome Archive (EGA) - for human genomic sequence data
- Federated European Genome Phenome Archive Sweden (FEGA Sweden) - Swedish EGA for human genomic sequence data
- ArrayExpress - for gene expression data
- PRIDE - for proteomics data
There are several ways to find domain-specific repositories within life science:
- EBI repository wizard - guide depending on data type
- ELIXIR deposition databases - core resources with long-term data preservation and accessibility plans
- FAIRsharing.org/databases - catalogue of many repositories, with possibility to filter on e.g. domain
- Scientific Data Repository Guidance - publisher’s recommendation
- re3data.org - registry of research data repositories (not only life science)
Repository metadata
The metadata you collect during your project will end up in the repository, hence finding a suitable repository already while planning the project may help you decide on which metadata to collect.
General purpose repositories
From time to time you will most likely come across situations when there is no suitable domain-specific repository for your data type, for example if you have a new data type. Another situation you might find yourself in is that you have done registry-based research with human data, and you are not allowed to share this data but still want to publish the methodology openly. Making a metadata record in a general purpose repository will then allow others to easily find it, without violating the agreements with the registry holders.
General purpose repositories usually accepts anything and everything related to research, i.e. they are also useful for other purposes, besides publishing research data, e.g. posters and presentations can be made publicly available and obtain a persistent identifier (DOI).
This type of repository is typically indexed, so you can find its content via search engines, and thus it has good reach. However, since the repository accepts many data types, the metadata will be less specific (no or very high level metadata standards), with the result that is more difficult for future users to judge if a dataset will be useful for them. The repositories usually have long-term sustainability plan, i.e. they will be available for a long time, but might cost (now or in future).
Examples:
The examples above is likely sufficient, but we recommend to also check if your institution have e.g. a figshare instance similar to the one SciLifeLab provides.
In-house/institutional repositories
Many institutions offer local repositories, and might even require that affiliated researchers put their research outputs there. However, unless the repository is indexed (so that search engines can find its content) this type of repository is for archive/backup purpose mainly.
You can also choose to create and host an in-house repository yourself, but that put a lot of responsibility on your shoulders. For how long will you be able to sustain it? It also requires considerable labour in order to make the repository FAIR, and without that the repository will have limited reach unless you also publish in a data catalogue.
Evaluate the suitability of a repository
How do you know if a repository is trustworthy? Say that you find a repository that might fit your purposes, how can you evaluate if it is suitable? Apart from accepting your type of data, there are some questions to consider when deciding if a certain repository is suitable or not:
- Are others in the community using it? Explore what datasets are already in it.
- While exploring it, is it easy to navigate / user-friendly?
- Is there support / guidance for submission and reuse?
- Is it sustainable, i.e. will the repository be around for a while? Is there a long-term plan for financing the repository, is it managed by a trustworthy group?
- Will the datasets obtain persistent identifiers? Is the repository itself FAIR?
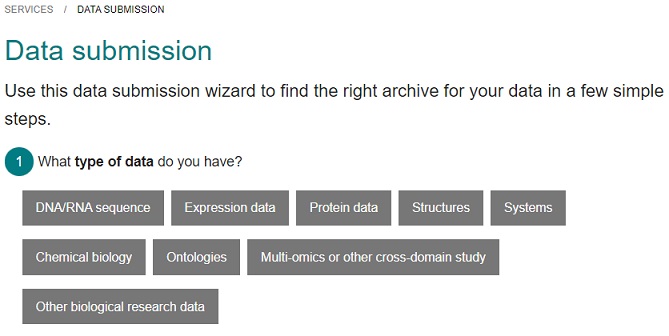
Demo of EBI Repository Wizard
EBI hosts several repositories, suitable for different types of life science data. The Repository Wizard helps you to identify which one is suitable for your data.
Go to the Wizard at https://www.ebi.ac.uk/submission/
Either explore the wizard with the purpose of finding a suitable repository for one of your projects, or choose among the scenarios provided below. Which repository is recommended?
- Genomics project with RNA sequences
- X-ray crystollography structure of a protein
- Gene expression data
- Protein sequencing data
- Proteomics project using mass spectrometry
- Electron microscopy structure images
Demo - Solution
- Genomics project with RNA sequences: European Nucleotide Archive (DNA/RNA sequence -> no controlled access -> produced experimentally -> Other)
- X-ray crystollography structure of a protein: wwPDB OneDep (Structures -> X-ray crystollography)
- Microarray gene expression data: ArrayExpress (Expression data -> no controlled access -> Microarray gene expression)
- Protein sequencing data: UniProt SPIN (Protein data -> no controlled access -> produced experimentally -> Protein sequencing)
- Proteomics project using mass spectrometry: PRIDE (Protein data -> no controlled access -> produced experimentally -> Mass spectrometry -> Proteomics)
- Electron microscopy structure images: EMPIAR (Structures-> Electron microscopy -> micrographs or particle stacks)