Scenario 5: Genetic Analysis Case Study on NRAS gene#
An Undiagnosed Child with Delayed Motor Development and Generalized Dysmorphism#
On clinical examination, the undiagnosed child was found to have several facial dysmorphisms, in particular a broad neck and low-set ears. It has also been assessed that the child exhibits delayed motor development.
Genetic testing has been performed using Whole Exome Sequencing (WES) which identified a probable pathogenic mutation in the NRAS gene.
Research Questions#
Question 1: Gene Mutation Prevalence#
Are there any other individuals with a mutation in the same gene?
To answer this question the SPARQL query below has been used for finding the phenopackets that include at least one mutation in the NRAS gene. To identify the correct gene descriptions, the HGNC gene ID is used of NRAS being HGNC:7989.
"PREFIX ex: <https://example.org/>\nPREFIX obo: <http://purl.obolibrary.org/obo/>\nPREFIX sio: <http://semanticscience.org/resource/>\nPREFIX dcterms: <http://purl.org/dc/terms/> \nPREFIX rdfs: <http://www.w3.org/2000/01/rdf-schema#>\n\nSELECT ?phenopacketidval ?creatorname ?createddate\nWHERE {\n # Find the phenopackets that contain at least one mutation in given gene\n ?genedescr a obo:NCIT_C16612 ;\n dcterms:identifier 'HGNC:7989' .\n ?vardescr sio:SIO_001403 ?genedescr .\n ?varinterpr sio:SIO_001403 ?vardescr .\n ?genomicinterp sio:SIO_001403 ?varinterpr .\n ?diagnosis sio:SIO_001403 ?genomicinterp .\n ?interpr sio:SIO_001403 ?diagnosis .\n ?phenopacket sio:SIO_001403 ?interpr ;\n a obo:NCIT_C79269 ;\n sio:SIO_000228 ?role .\n \n # Find phenopacket ID value\n ?phenopacketid sio:SIO_000020 ?role ;\n\t sio:SIO_000300 ?phenopacketidval .\n \n # Find metadata\n ?phenopacket sio:SIO_001403 ?metadata .\n ?metadata a obo:NCIT_C52095 ;\n sio:SIO_000008 ?creator ;\n sio:SIO_000008 ?created .\n \n ?creator a dcterms:creator .\n ?creatorval sio:SIO_000628 ?creator ;\n a obo:NCIT_C70856 ;\n sio:SIO_000300 ?creatorname .\n \n ?created a dcterms:created .\n ?createdval sio:SIO_000628 ?created ;\n a obo:NCIT_C70856 ;\n sio:SIO_000300 ?createddate .\n}"
As shown below, a total of fourteen phenopackets have been found including phenotypic and genetic information related to mutations in the NRAS gene. This set of phenopackets has been created by the same researcher at the same datetime based on the metadata.
| phenopacket id | creator | creation date | |
|---|---|---|---|
| 0 | PMID_26467218_individual_6_Cirstea_et_al__14 | ORCID:0000-0002-0736-9199 | 2024-05-20 20:56:12 |
| 1 | PMID_26467218_individual_11_Kraoua_et_al__23 | ORCID:0000-0002-0736-9199 | 2024-05-20 20:56:12 |
| 2 | PMID_26467218_individual_3_Denayer_et_al__22 | ORCID:0000-0002-0736-9199 | 2024-05-20 20:56:12 |
| 3 | PMID_26467218_individual_9_Cirstea_et_al__14 | ORCID:0000-0002-0736-9199 | 2024-05-20 20:56:12 |
| 4 | PMID_26467218_individual_4_Denayer_et_al__22 | ORCID:0000-0002-0736-9199 | 2024-05-20 20:56:12 |
| 5 | PMID_26467218_individual_12_Present_study | ORCID:0000-0002-0736-9199 | 2024-05-20 20:56:12 |
| 6 | PMID_26467218_individual_1_De_Filippi_et_al__20 | ORCID:0000-0002-0736-9199 | 2024-05-20 20:56:12 |
| 7 | PMID_26467218_individual_2_Runtuwene_et_al__21 | ORCID:0000-0002-0736-9199 | 2024-05-20 20:56:12 |
| 8 | PMID_26467218_individual_9_M_Cirstea_et_al__14 | ORCID:0000-0002-0736-9199 | 2024-05-20 20:56:12 |
| 9 | PMID_26467218_individual_5_Denayer_et_al__22 | ORCID:0000-0002-0736-9199 | 2024-05-20 20:56:12 |
| 10 | PMID_26467218_individual_12_F_Present_study | ORCID:0000-0002-0736-9199 | 2024-05-20 20:56:12 |
| 11 | PMID_26467218_individual_8_Cirstea_et_al__14 | ORCID:0000-0002-0736-9199 | 2024-05-20 20:56:12 |
| 12 | PMID_26467218_individual_10_Kraoua_et_al__23 | ORCID:0000-0002-0736-9199 | 2024-05-20 20:56:12 |
| 13 | PMID_26467218_individual_7_Cirstea_et_al__14 | ORCID:0000-0002-0736-9199 | 2024-05-20 20:56:12 |
Question 2: Phenotype and Disease Prevalence#
What phenotypes and which diseases have been observed in these individuals and how often do they occur in this group of individuals?
The phenopackets found for the previous question can now be analysed to conclude which phenotypes and diseases occur in this group of individuals. This is done by using SPARQL queries for acquiring the associated phenotypes and diseases. The query below searches for all diagnosed diseases in the relevant phenopackets.
'PREFIX ex: <https://example.org/>\nPREFIX obo: <http://purl.obolibrary.org/obo/>\nPREFIX sio: <http://semanticscience.org/resource/>\nPREFIX dcterms: <http://purl.org/dc/terms/> \nPREFIX rdfs: <http://www.w3.org/2000/01/rdf-schema#>\n\nSELECT ?idvalue ?diseaseid ?diseaselabel\nWHERE {\n # Find relevant phenopackets\n ?id a obo:IAO_0020000 ;\n sio:SIO_000300 ?idvalue .\n VALUES ?idvalue { "PMID_26467218_individual_6_Cirstea_et_al__14" "PMID_26467218_individual_11_Kraoua_et_al__23" "PMID_26467218_individual_3_Denayer_et_al__22" "PMID_26467218_individual_9_Cirstea_et_al__14" "PMID_26467218_individual_4_Denayer_et_al__22" "PMID_26467218_individual_12_Present_study" "PMID_26467218_individual_1_De_Filippi_et_al__20" "PMID_26467218_individual_2_Runtuwene_et_al__21" "PMID_26467218_individual_9_M_Cirstea_et_al__14" "PMID_26467218_individual_5_Denayer_et_al__22" "PMID_26467218_individual_12_F_Present_study" "PMID_26467218_individual_8_Cirstea_et_al__14" "PMID_26467218_individual_10_Kraoua_et_al__23" "PMID_26467218_individual_7_Cirstea_et_al__14" } .\n \n # Find diagnosis\n ?id sio:SIO_000020 ?role .\n ?phenopacket sio:SIO_000228 ?role ;\n sio:SIO_001403 ?interpr .\n ?interpr a obo:NCIT_C41255 ;\n sio:SIO_001403 ?diagnosis .\n ?diagnosis a sio:SIO_000614 ;\n sio:SIO_001403 ?disease .\n ?diseaseval a obo:NCIT_C2991 ;\n sio:SIO_000628 ?disease .\n ?disease rdfs:label ?diseaselabel ;\n dcterms:identifier ?diseaseid .\n}'
Given the results of the SPARQL query shown above, the individuals for which mutations are found in the NRAS gene are all diagnosed with Noonan syndrome 6.
| phenopacket id | disease ID | disease label | |
|---|---|---|---|
| 0 | PMID_26467218_individual_6_Cirstea_et_al__14 | OMIM:613224 | Noonan syndrome 6 |
| 1 | PMID_26467218_individual_11_Kraoua_et_al__23 | OMIM:613224 | Noonan syndrome 6 |
| 2 | PMID_26467218_individual_3_Denayer_et_al__22 | OMIM:613224 | Noonan syndrome 6 |
| 3 | PMID_26467218_individual_9_Cirstea_et_al__14 | OMIM:613224 | Noonan syndrome 6 |
| 4 | PMID_26467218_individual_4_Denayer_et_al__22 | OMIM:613224 | Noonan syndrome 6 |
| 5 | PMID_26467218_individual_12_Present_study | OMIM:613224 | Noonan syndrome 6 |
| 6 | PMID_26467218_individual_1_De_Filippi_et_al__20 | OMIM:613224 | Noonan syndrome 6 |
| 7 | PMID_26467218_individual_2_Runtuwene_et_al__21 | OMIM:613224 | Noonan syndrome 6 |
| 8 | PMID_26467218_individual_9_M_Cirstea_et_al__14 | OMIM:613224 | Noonan syndrome 6 |
| 9 | PMID_26467218_individual_5_Denayer_et_al__22 | OMIM:613224 | Noonan syndrome 6 |
| 10 | PMID_26467218_individual_12_F_Present_study | OMIM:613224 | Noonan syndrome 6 |
| 11 | PMID_26467218_individual_8_Cirstea_et_al__14 | OMIM:613224 | Noonan syndrome 6 |
| 12 | PMID_26467218_individual_10_Kraoua_et_al__23 | OMIM:613224 | Noonan syndrome 6 |
| 13 | PMID_26467218_individual_7_Cirstea_et_al__14 | OMIM:613224 | Noonan syndrome 6 |
The query shown below searches for all phenotypes that are associated with at least one of the relevant phenopackets.
'PREFIX ex: <https://example.org/>\nPREFIX obo: <http://purl.obolibrary.org/obo/>\nPREFIX sio: <http://semanticscience.org/resource/>\nPREFIX dcterms: <http://purl.org/dc/terms/> \nPREFIX rdfs: <http://www.w3.org/2000/01/rdf-schema#>\n\nSELECT ?idvalue ?phenofeatid ?phenofeatlabel\nWHERE {\n # Find relevant phenopackets\n ?id a obo:IAO_0020000 ;\n sio:SIO_000300 ?idvalue .\n VALUES ?idvalue { "PMID_26467218_individual_6_Cirstea_et_al__14" "PMID_26467218_individual_11_Kraoua_et_al__23" "PMID_26467218_individual_3_Denayer_et_al__22" "PMID_26467218_individual_9_Cirstea_et_al__14" "PMID_26467218_individual_4_Denayer_et_al__22" "PMID_26467218_individual_12_Present_study" "PMID_26467218_individual_1_De_Filippi_et_al__20" "PMID_26467218_individual_2_Runtuwene_et_al__21" "PMID_26467218_individual_9_M_Cirstea_et_al__14" "PMID_26467218_individual_5_Denayer_et_al__22" "PMID_26467218_individual_12_F_Present_study" "PMID_26467218_individual_8_Cirstea_et_al__14" "PMID_26467218_individual_10_Kraoua_et_al__23" "PMID_26467218_individual_7_Cirstea_et_al__14" } .\n \n # Find phenotypic features\n ?id sio:SIO_000020 ?role .\n ?phenopacket sio:SIO_000228 ?role ;\n sio:SIO_001403 ?phenofeat .\n ?phenofeatval sio:SIO_000628 ?phenofeat ;\n a obo:NCIT_C16977 .\n ?phenofeat dcterms:identifier ?phenofeatid ;\n rdfs:label ?phenofeatlabel .\n \n}'
A total of 326 phenotypes have been found. A sample of these results are shown below:
| phenopacket ID | phenotype ID | phenotype label | |
|---|---|---|---|
| 94 | PMID_26467218_individual_9_Cirstea_et_al__14 | HP:0000028 | Cryptorchidism |
| 133 | PMID_26467218_individual_12_Present_study | HP:0000767 | Pectus excavatum |
| 179 | PMID_26467218_individual_2_Runtuwene_et_al__21 | HP:0000256 | Macrocephaly |
| 184 | PMID_26467218_individual_9_M_Cirstea_et_al__14 | HP:0000508 | Ptosis |
| 198 | PMID_26467218_individual_9_M_Cirstea_et_al__14 | HP:0006699 | Premature atrial contractions |
| 207 | PMID_26467218_individual_5_Denayer_et_al__22 | HP:0000256 | Macrocephaly |
| 233 | PMID_26467218_individual_12_F_Present_study | HP:0001561 | Polyhydramnios |
| 292 | PMID_26467218_individual_10_Kraoua_et_al__23 | HP:0001680 | Coarctation of aorta |
| 324 | PMID_26467218_individual_7_Cirstea_et_al__14 | HP:0006699 | Premature atrial contractions |
| 325 | PMID_26467218_individual_7_Cirstea_et_al__14 | HP:0002212 | Curly hair |
Given these results, the occurrence of each phenotype is calculated:
| phenotype ID | phenotype label | counts | occurrence percentage | |
|---|---|---|---|---|
| 0 | HP:0000369 | Low-set ears | 14 | 100.0% |
| 1 | HP:0000316 | Hypertelorism | 14 | 100.0% |
| 2 | HP:0000957 | Cafe-au-lait spot | 14 | 100.0% |
| 3 | HP:0012209 | Juvenile myelomonocytic leukemia | 14 | 100.0% |
| 4 | HP:0000494 | Downslanted palpebral fissures | 14 | 100.0% |
| 5 | HP:0004322 | Short stature | 14 | 100.0% |
| 6 | HP:0000348 | High forehead | 14 | 100.0% |
| 7 | HP:0000508 | Ptosis | 14 | 100.0% |
| 8 | HP:0000358 | Posteriorly rotated ears | 14 | 100.0% |
| 9 | HP:0000767 | Pectus excavatum | 13 | 92.9% |
| 10 | HP:0001655 | Patent foramen ovale | 13 | 92.9% |
| 11 | HP:0000256 | Macrocephaly | 13 | 92.9% |
| 12 | HP:0001680 | Coarctation of aorta | 13 | 92.9% |
| 13 | HP:0000483 | Astigmatism | 12 | 85.7% |
| 14 | HP:0001270 | Motor delay | 12 | 85.7% |
| 15 | HP:0000486 | Strabismus | 12 | 85.7% |
| 16 | HP:0000465 | Webbed neck | 12 | 85.7% |
| 17 | HP:0000545 | Myopia | 11 | 78.6% |
| 18 | HP:0000563 | Keratoconus | 11 | 78.6% |
| 19 | HP:0006699 | Premature atrial contractions | 11 | 78.6% |
| 20 | HP:0002212 | Curly hair | 11 | 78.6% |
| 21 | HP:0000978 | Bruising susceptibility | 11 | 78.6% |
| 22 | HP:0001561 | Polyhydramnios | 10 | 71.4% |
| 23 | HP:0001195 | Single umbilical artery | 10 | 71.4% |
| 24 | HP:0032152 | Keratosis pilaris | 9 | 64.3% |
| 25 | HP:0000028 | Cryptorchidism | 9 | 64.3% |
| 26 | HP:0001639 | Hypertrophic cardiomyopathy | 3 | 21.4% |
| 27 | HP:0001642 | Pulmonic stenosis | 3 | 21.4% |
| 28 | HP:0001631 | Atrial septal defect | 1 | 7.1% |
Question 3: Matching Phenotypes#
Do the found phenotypes overlap with the phenotypes observed in the undiagnosed child?
Multiple phenotypes have been observed in the undiagnosed child being broad neck, low-set ears and motor delay. In order to match the observed phenotypes with the phenotypes observed in the phenopackets, the Human Phenotype Ontology (HPO) identifiers need to be retrieved. These identifiers are acquired by querying over this HPO ontology matching the phenotypes with the labels of the terms that are part of HPO. Below, a SPARQL query is shown that searches for the identifier for the broad neck phenotype:
"PREFIX rdfs: <http://www.w3.org/2000/01/rdf-schema#>\nPREFIX obo: <http://purl.obolibrary.org/obo/>\n\nSELECT ?iri\nWHERE {\n ?iri rdfs:label 'Broad neck' .\n}"
Executing this SPARQL query for all observed phenotypes results in the following identifiers:
| id | label | |
|---|---|---|
| 0 | HP:0000475 | Broad neck |
| 1 | HP:0000369 | Low-set ears |
| 2 | HP:0001270 | Motor delay |
Next, the list of observed phenotypes can be compared with the list of phenotypes associated with the selection of phenopackets coming to the conclusion that the observed phenotypes Motor delay (HP:0001270), Low-set ears (HP:0000369) are also found in the individuals represented in the phenopackets.
It is still possible to gather more information about the observed phenotype that has not been found in at least one of the phenopackets. One step would be to investigate the closeness of the observed phenotype to the phenotypes associated with Noonan syndrome 6 given their relative positions in the HPO ontology. To be more explicit, this closeness can be calculated by acquiring the shortest path length between phenotypes in a network. This network is built by adding each phenotype as a node. An edge between two nodes represents the relation of one phenotype being the subclass or superclass of another phenotype.
Given the path length distances between each pair of phenotypes, the phenotypes can be shown on a two-dimensional plot by applying multidimensional scaling. In this way, a clear overview is generated of all phenotypes and their similarities expressed in their positions in the two dimensional space. This plot is shown below:
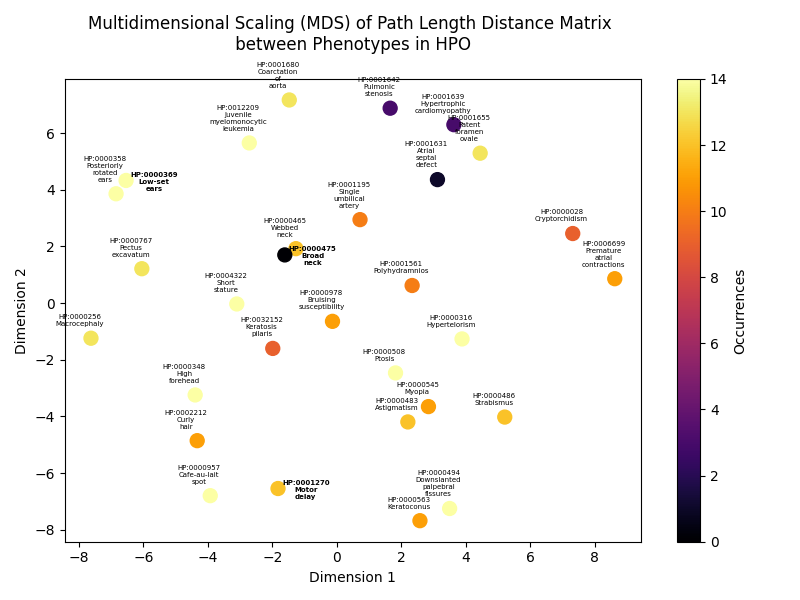
The phenotypes labeled in bold text are the phenotypes observed in the undiagnosed child. Again, it can be seen that the phenotypes low-set ears and motor delay are also found in patients with Noonan syndrome 6 given the colors of the points that represent the phenotypes. Interestingly, this plot also shows that the remaining observed phenotype “broad neck” seems to be similar to phenotype “webbed neck” that is also associated in group of individuals with this same diagnosis.
Conclusion#
Given the mutations in the NRAS gene identified in individuals with Noonan syndrome 6 and the overlap and similarities between the observed phenotypes and the phenotypes associated with the individuals diagnosed with Noonan syndrome 6, there is reason to further investigate Noonan syndrome 6 as being a likely diagnosis for the child.
