class: center, middle, inverse, title-slide # Working with <code>ggplot2</code> ## RaukR 2019 • Advanced R for Bioinformatics ### <b>Roy Francis</b> ### NBIS, SciLifeLab --- exclude: true count: false <link href="https://fonts.googleapis.com/css?family=Roboto|Source+Sans+Pro:300,400,600|Ubuntu+Mono&subset=latin-ext" rel="stylesheet"> <!-- ----------------- Only edit title & author above this ----------------- --> ## Contents * [Why `ggplot2`?](#intro) * [Grammar of Graphics](#gog) * [Data](#data-iris) * [Geoms](#geom) * [Stats](#stat-1) * [Aesthetics](#aes) * [Scales](#scales-discrete-colour) * [Facets](#facet-wrap) * [Coordinates](#coordinate) * [Theme](#theme) * [Position](#position) * [Saving Plots](#save) * [Combining Plots](#comb) * [Interactive Plots](#interactive) * [Extensions](#extension) --- name: intro class: spaced ## Why `ggplot2`? * Consistent code * Flexible * Automatic legends, colors etc * Save plot objects * Themes for reusing styles * Numerous add-ons/extensions * Nearly complete graphing solution -- Not suitable for: * 3D graphics ??? Why can't we just do everything is base plot? Of course, we could, but it's easier, consistent and more structured using `ggplot2`. There is bit of a learning curve, but once the code syntax and graphic building logic is clear, it becomes easy to plot a large variety of graphs. --- name: gvb1 ## `ggplot2` vs Base Graphics .pull-left-50[ ```r hist(iris$Sepal.Length) ``` <img src="ggplot_presentation_files/figure-html/unnamed-chunk-3-1.svg" style="display: block; margin: auto auto auto 0;" /> ] .pull-right-50[ ```r library(ggplot2) ggplot(iris,aes(x=Sepal.Length))+ geom_histogram(bins=8) ``` <img src="ggplot_presentation_files/figure-html/unnamed-chunk-4-1.svg" style="display: block; margin: auto auto auto 0;" /> ] ??? For simple graphs, the base plot seem to take minimal coding effort compared to a ggplot graph. --- name: gvb2 ## `ggplot2` vs Base Graphics .pull-left-50[ ```r plot(iris$Petal.Length,iris$Petal.Width, col=c("red","green","blue")[iris$Species], pch=c(0,1,2)[iris$Species]) legend(x=1,y=2.5, legend=c("setosa","versicolor","virginica"), pch=c(0,1,2),col=c("red","green","blue")) ``` <img src="ggplot_presentation_files/figure-html/unnamed-chunk-5-1.svg" style="display: block; margin: auto auto auto 0;" /> ] .pull-right-50[ ```r ggplot(iris,aes(Petal.Length,Sepal.Length,color=Species))+ geom_point() ``` <img src="ggplot_presentation_files/figure-html/unnamed-chunk-6-1.svg" style="display: block; margin: auto auto auto 0;" /> ] ??? For anything beyond extremely basic plots, base plotting quickly become complex. More importantly, base plots do not have consistency in it's functions or plotting strategy. --- name: gog class: spaced ## Grammar Of Graphics .pull-left-30[  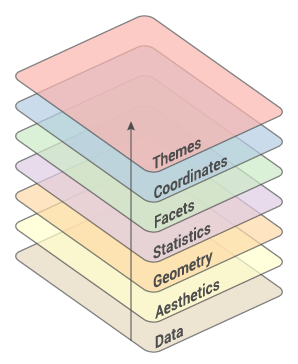 ] -- .pull-right-70[ * **Data**: Input data * **Geom**: A geometry representing data. Points, Lines etc * **Aesthetic**: Visual characteristics of the geometry. Size, Color, Shape etc * **Scale**: How visual characteristics are converted to display values * **Statistics**: Statistical transformations. Counts, Means etc * **Coordinates**: Numeric system to determine position of geometry. Cartesian, Polar etc * **Facets**: Split data into subsets ] ??? `ggplot` was created by Hadley Wickham in 2005 as an implementation of Leland Wilkinson's book Grammar of Graphics. Different graphs have always been considered as independent entities and also labelled differently such as barplots, scatterplots, boxplots etc. Each graph has it's own function and plotting strategy. Grammar of graphics (GOG) tries to unify all graphs under a common umbrella. GOG brings the idea that graphs are made up of discrete components which can be mixed and matched to create any plot. This creates a consistent underlying framework to graphing. --- name: syntax ## Building A Graph: Syntax  --- name: build-1 ## Building A Graph .pull-left-40[ ```r ggplot(iris) ``` ] .pull-right-50[ <img src="ggplot_presentation_files/figure-html/unnamed-chunk-8-1.svg" style="display: block; margin: auto auto auto 0;" /> ] --- name: build-2 ## Building A Graph .pull-left-40[ ```r ggplot(iris,aes(x=Sepal.Length, y=Sepal.Width)) ``` ] .pull-right-50[ <img src="ggplot_presentation_files/figure-html/unnamed-chunk-10-1.svg" style="display: block; margin: auto auto auto 0;" /> ] --- name: build-3 ## Building A Graph .pull-left-40[ ```r ggplot(iris,aes(x=Sepal.Length, y=Sepal.Width))+ geom_point() ``` ] .pull-right-50[ <img src="ggplot_presentation_files/figure-html/unnamed-chunk-12-1.svg" style="display: block; margin: auto auto auto 0;" /> ] --- name: build-4 ## Building A Graph .pull-left-40[ ```r ggplot(iris,aes(x=Sepal.Length, y=Sepal.Width, colour=Species))+ geom_point() ``` ] .pull-right-50[ <img src="ggplot_presentation_files/figure-html/unnamed-chunk-14-1.svg" style="display: block; margin: auto auto auto 0;" /> ] --- name: data-iris ## Data • `iris` * Input data is always an R `data.frame` object <table class="table table-striped table-hover table-responsive table-condensed" style="width: auto !important; "> <thead> <tr> <th style="text-align:center;"> Sepal.Length </th> <th style="text-align:center;"> Sepal.Width </th> <th style="text-align:center;"> Petal.Length </th> <th style="text-align:center;"> Petal.Width </th> <th style="text-align:center;"> Species </th> </tr> </thead> <tbody> <tr> <td style="text-align:center;"> 5.1 </td> <td style="text-align:center;"> 3.5 </td> <td style="text-align:center;"> 1.4 </td> <td style="text-align:center;"> 0.2 </td> <td style="text-align:center;"> setosa </td> </tr> <tr> <td style="text-align:center;"> 4.9 </td> <td style="text-align:center;"> 3.0 </td> <td style="text-align:center;"> 1.4 </td> <td style="text-align:center;"> 0.2 </td> <td style="text-align:center;"> setosa </td> </tr> <tr> <td style="text-align:center;"> 4.7 </td> <td style="text-align:center;"> 3.2 </td> <td style="text-align:center;"> 1.3 </td> <td style="text-align:center;"> 0.2 </td> <td style="text-align:center;"> setosa </td> </tr> <tr> <td style="text-align:center;"> 4.6 </td> <td style="text-align:center;"> 3.1 </td> <td style="text-align:center;"> 1.5 </td> <td style="text-align:center;"> 0.2 </td> <td style="text-align:center;"> setosa </td> </tr> <tr> <td style="text-align:center;"> 5.0 </td> <td style="text-align:center;"> 3.6 </td> <td style="text-align:center;"> 1.4 </td> <td style="text-align:center;"> 0.2 </td> <td style="text-align:center;"> setosa </td> </tr> <tr> <td style="text-align:center;"> 5.4 </td> <td style="text-align:center;"> 3.9 </td> <td style="text-align:center;"> 1.7 </td> <td style="text-align:center;"> 0.4 </td> <td style="text-align:center;"> setosa </td> </tr> </tbody> </table> ```r str(iris) ``` ``` ## 'data.frame': 150 obs. of 5 variables: ## $ Sepal.Length: num 5.1 4.9 4.7 4.6 5 5.4 4.6 5 4.4 4.9 ... ## $ Sepal.Width : num 3.5 3 3.2 3.1 3.6 3.9 3.4 3.4 2.9 3.1 ... ## $ Petal.Length: num 1.4 1.4 1.3 1.5 1.4 1.7 1.4 1.5 1.4 1.5 ... ## $ Petal.Width : num 0.2 0.2 0.2 0.2 0.2 0.4 0.3 0.2 0.2 0.1 ... ## $ Species : Factor w/ 3 levels "setosa","versicolor",..: 1 1 1 1 1 1 1 1 1 1 ... ``` ??? It's a good idea to use `str()` to check the input dataframe to make sure that numbers are actually numbers and not characters, for example. Verify that factors are correctly assigned. --- name: data-diamonds ## Data • `diamonds` <table class="table table-striped table-hover table-responsive table-condensed" style="width: auto !important; "> <thead> <tr> <th style="text-align:center;"> carat </th> <th style="text-align:center;"> cut </th> <th style="text-align:center;"> color </th> <th style="text-align:center;"> clarity </th> <th style="text-align:center;"> depth </th> <th style="text-align:center;"> table </th> <th style="text-align:center;"> price </th> <th style="text-align:center;"> x </th> <th style="text-align:center;"> y </th> <th style="text-align:center;"> z </th> </tr> </thead> <tbody> <tr> <td style="text-align:center;"> 0.23 </td> <td style="text-align:center;"> Ideal </td> <td style="text-align:center;"> E </td> <td style="text-align:center;"> SI2 </td> <td style="text-align:center;"> 61.5 </td> <td style="text-align:center;"> 55 </td> <td style="text-align:center;"> 326 </td> <td style="text-align:center;"> 3.95 </td> <td style="text-align:center;"> 3.98 </td> <td style="text-align:center;"> 2.43 </td> </tr> <tr> <td style="text-align:center;"> 0.21 </td> <td style="text-align:center;"> Premium </td> <td style="text-align:center;"> E </td> <td style="text-align:center;"> SI1 </td> <td style="text-align:center;"> 59.8 </td> <td style="text-align:center;"> 61 </td> <td style="text-align:center;"> 326 </td> <td style="text-align:center;"> 3.89 </td> <td style="text-align:center;"> 3.84 </td> <td style="text-align:center;"> 2.31 </td> </tr> <tr> <td style="text-align:center;"> 0.23 </td> <td style="text-align:center;"> Good </td> <td style="text-align:center;"> E </td> <td style="text-align:center;"> VS1 </td> <td style="text-align:center;"> 56.9 </td> <td style="text-align:center;"> 65 </td> <td style="text-align:center;"> 327 </td> <td style="text-align:center;"> 4.05 </td> <td style="text-align:center;"> 4.07 </td> <td style="text-align:center;"> 2.31 </td> </tr> <tr> <td style="text-align:center;"> 0.29 </td> <td style="text-align:center;"> Premium </td> <td style="text-align:center;"> I </td> <td style="text-align:center;"> VS2 </td> <td style="text-align:center;"> 62.4 </td> <td style="text-align:center;"> 58 </td> <td style="text-align:center;"> 334 </td> <td style="text-align:center;"> 4.20 </td> <td style="text-align:center;"> 4.23 </td> <td style="text-align:center;"> 2.63 </td> </tr> <tr> <td style="text-align:center;"> 0.31 </td> <td style="text-align:center;"> Good </td> <td style="text-align:center;"> J </td> <td style="text-align:center;"> SI2 </td> <td style="text-align:center;"> 63.3 </td> <td style="text-align:center;"> 58 </td> <td style="text-align:center;"> 335 </td> <td style="text-align:center;"> 4.34 </td> <td style="text-align:center;"> 4.35 </td> <td style="text-align:center;"> 2.75 </td> </tr> <tr> <td style="text-align:center;"> 0.24 </td> <td style="text-align:center;"> Very Good </td> <td style="text-align:center;"> J </td> <td style="text-align:center;"> VVS2 </td> <td style="text-align:center;"> 62.8 </td> <td style="text-align:center;"> 57 </td> <td style="text-align:center;"> 336 </td> <td style="text-align:center;"> 3.94 </td> <td style="text-align:center;"> 3.96 </td> <td style="text-align:center;"> 2.48 </td> </tr> </tbody> </table> ```r str(diamonds) ``` ``` ## tibble [53,940 × 10] (S3: tbl_df/tbl/data.frame) ## $ carat : num [1:53940] 0.23 0.21 0.23 0.29 0.31 0.24 0.24 0.26 0.22 0.23 ... ## $ cut : Ord.factor w/ 5 levels "Fair"<"Good"<..: 5 4 2 4 2 3 3 3 1 3 ... ## $ color : Ord.factor w/ 7 levels "D"<"E"<"F"<"G"<..: 2 2 2 6 7 7 6 5 2 5 ... ## $ clarity: Ord.factor w/ 8 levels "I1"<"SI2"<"SI1"<..: 2 3 5 4 2 6 7 3 4 5 ... ## $ depth : num [1:53940] 61.5 59.8 56.9 62.4 63.3 62.8 62.3 61.9 65.1 59.4 ... ## $ table : num [1:53940] 55 61 65 58 58 57 57 55 61 61 ... ## $ price : int [1:53940] 326 326 327 334 335 336 336 337 337 338 ... ## $ x : num [1:53940] 3.95 3.89 4.05 4.2 4.34 3.94 3.95 4.07 3.87 4 ... ## $ y : num [1:53940] 3.98 3.84 4.07 4.23 4.35 3.96 3.98 4.11 3.78 4.05 ... ## $ z : num [1:53940] 2.43 2.31 2.31 2.63 2.75 2.48 2.47 2.53 2.49 2.39 ... ``` ??? R `data.frame` is a tabular format with rows and columns just like a spreadsheet. All items in a row or a column must be available or missing values filled in as NAs. --- name: data-format ## Data • Format .size-80[] **Wide** <table class="table table-striped table-hover table-responsive table-condensed" style="width: auto !important; "> <thead> <tr> <th style="text-align:center;"> Sepal.Length </th> <th style="text-align:center;"> Sepal.Width </th> <th style="text-align:center;"> Petal.Length </th> <th style="text-align:center;"> Petal.Width </th> <th style="text-align:center;"> Species </th> </tr> </thead> <tbody> <tr> <td style="text-align:center;"> 5.1 </td> <td style="text-align:center;"> 3.5 </td> <td style="text-align:center;"> 1.4 </td> <td style="text-align:center;"> 0.2 </td> <td style="text-align:center;"> setosa </td> </tr> <tr> <td style="text-align:center;"> 4.9 </td> <td style="text-align:center;"> 3.0 </td> <td style="text-align:center;"> 1.4 </td> <td style="text-align:center;"> 0.2 </td> <td style="text-align:center;"> setosa </td> </tr> <tr> <td style="text-align:center;"> 4.7 </td> <td style="text-align:center;"> 3.2 </td> <td style="text-align:center;"> 1.3 </td> <td style="text-align:center;"> 0.2 </td> <td style="text-align:center;"> setosa </td> </tr> </tbody> </table> **Long** <table class="table table-striped table-hover table-responsive table-condensed" style="width: auto !important; "> <thead> <tr> <th style="text-align:center;"> Species </th> <th style="text-align:center;"> variable </th> <th style="text-align:center;"> value </th> </tr> </thead> <tbody> <tr> <td style="text-align:center;"> setosa </td> <td style="text-align:center;"> Sepal.Length </td> <td style="text-align:center;"> 5.1 </td> </tr> <tr> <td style="text-align:center;"> setosa </td> <td style="text-align:center;"> Sepal.Length </td> <td style="text-align:center;"> 4.9 </td> </tr> <tr> <td style="text-align:center;"> setosa </td> <td style="text-align:center;"> Sepal.Length </td> <td style="text-align:center;"> 4.7 </td> </tr> </tbody> </table> ??? The data must be cleaned up and prepared for plotting. The data must be 'tidy'. Columns must be variables and rows must be observations. The data can then be in wide or long format depending on the variables to be plotted. --- name: geom ## Geoms 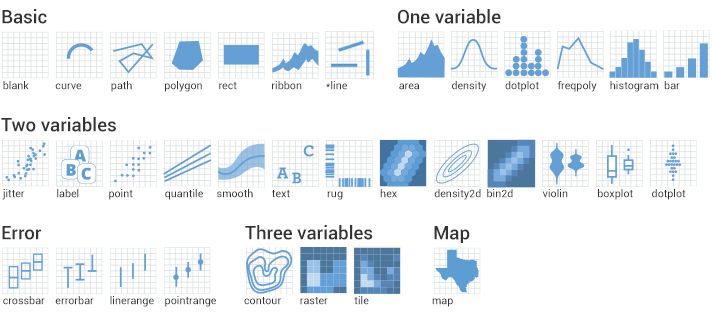 -- ```r p <- ggplot(iris) # scatterplot p+geom_point(aes(x=Sepal.Length,y=Sepal.Width)) # barplot p+geom_bar(aes(x=Sepal.Length)) # boxplot p+geom_boxplot(aes(x=Species,y=Sepal.Width)) # search help.search("^geom_",package="ggplot2") ``` ??? Geoms are the geometric components of a graph such as points, lines etc used to represent data. The same data can be visually represented in different geoms. For example, points or bars. Mandatory input requirements change depending on geoms. --- name: stat-1 ## Stats * Stats compute new variables from input data. -- * Geoms have default stats. ```r x <- ggplot(iris) + geom_bar(aes(x=Sepal.Length),stat="bin") y <- ggplot(iris) + geom_bar(aes(x=Species),stat="count") z <- ggplot(iris) + geom_bar(aes(x=Species,y=Sepal.Length),stat="identity") grid.arrange(x,y,z,nrow=1) ``` <img src="ggplot_presentation_files/figure-html/unnamed-chunk-22-1.svg" style="display: block; margin: auto auto auto 0;" /> -- * Plots can be built with stats. ```r x <- ggplot(iris) + stat_bin(aes(x=Sepal.Length),geom="bar") y <- ggplot(iris) + stat_count(aes(x=Species),geom="bar") z <- ggplot(iris) + stat_identity(aes(x=Species,y=Sepal.Length),geom="bar") grid.arrange(x,y,z,nrow=1) ``` <img src="ggplot_presentation_files/figure-html/unnamed-chunk-23-1.svg" style="display: block; margin: auto auto auto 0;" /> ??? * Normally the data is plotted directly from input as it is. * Some plots require the data to be computed or transformed. Eg. boxplot, histograms, smoothing, predictions, regression etc. --- name: stat-2 ## Stats * Stats have default geoms. <table class="table table-striped table-hover table-responsive table-condensed" style="width: auto !important; "> <thead> <tr> <th style="text-align:left;"> plot </th> <th style="text-align:left;"> stat </th> <th style="text-align:left;"> geom </th> </tr> </thead> <tbody> <tr> <td style="text-align:left;"> histogram </td> <td style="text-align:left;"> bin </td> <td style="text-align:left;"> bar </td> </tr> <tr> <td style="text-align:left;"> smooth </td> <td style="text-align:left;"> smooth </td> <td style="text-align:left;"> line </td> </tr> <tr> <td style="text-align:left;"> boxplot </td> <td style="text-align:left;"> boxplot </td> <td style="text-align:left;"> boxplot </td> </tr> <tr> <td style="text-align:left;"> density </td> <td style="text-align:left;"> density </td> <td style="text-align:left;"> line </td> </tr> <tr> <td style="text-align:left;"> freqpoly </td> <td style="text-align:left;"> freqpoly </td> <td style="text-align:left;"> line </td> </tr> </tbody> </table> Use `args(geom_bar)` to check arguments. --- name: aes ## Aesthetics * Aesthetic mapping vs aesthetic parameter .pull-left-50[ ```r ggplot(iris)+ geom_point(aes(x=Sepal.Length, y=Sepal.Width, size=Petal.Length, alpha=Petal.Width, shape=Species, color=Species)) ``` <img src="ggplot_presentation_files/figure-html/unnamed-chunk-26-1.svg" style="display: block; margin: auto auto auto 0;" /> ] .pull-left-50[ ```r ggplot(iris)+ geom_point(aes(x=Sepal.Length, y=Sepal.Width), size=2, alpha=0.8, shape=15, color="steelblue") ``` <img src="ggplot_presentation_files/figure-html/unnamed-chunk-27-1.svg" style="display: block; margin: auto auto auto 0;" /> ] ??? Aesthetics are used to assign values to geometries. For example, a set of points can be a fixed size or can be different colors or sizes denoting a variable. This would be an incorrect way to do it. ``` ggplot(iris)+ geom_point(aes(x=Sepal.Length,y=Sepal.Width,size=2) ``` --- name: aes-2 ## Aesthetics ```r x1 <- ggplot(iris) + geom_point(aes(x=Sepal.Length,y=Sepal.Width))+ stat_smooth(aes(x=Sepal.Length,y=Sepal.Width)) x2 <- ggplot(iris,aes(x=Sepal.Length,y=Sepal.Width))+ geom_point() + geom_smooth() grid.arrange(x1,x2,nrow=1,ncol=2) ``` <img src="ggplot_presentation_files/figure-html/unnamed-chunk-28-1.svg" style="display: block; margin: auto auto auto 0;" /> ??? If the same aesthetics are used in multiple geoms, they can be moved to `ggplot()`. --- name: multiple-geom ## Multiple Geoms ```r ggplot(iris,aes(x=Sepal.Length,y=Sepal.Width))+ geom_point()+ geom_line()+ geom_smooth()+ geom_rug()+ geom_step()+ geom_text(data=subset(iris,iris$Species=="setosa"),aes(label=Species)) ``` <img src="ggplot_presentation_files/figure-html/unnamed-chunk-29-1.svg" style="display: block; margin: auto auto auto 0;" /> ??? Multiple geoms can be plotted one after the other. The order in which items are specified in the command dictates the plotting order on the actual plot. In this case, the points appear over the lines. ``` ggplot(iris,aes(x=Sepal.Length,y=Sepal.Width))+ geom_point()+ geom_line()+ ``` while here the lines appear above the points. ``` ggplot(iris,aes(x=Sepal.Length,y=Sepal.Width))+ geom_line()+ geom_point()+ ``` Each geom takes input from `ggplot()` inputs. If extra input is required to a geom, it can be specified additionally inside `aes()`. `data` can be changed if needed for specific geoms. --- name: scales-discrete-color ## Scales • Discrete Colors * scales: position, color, fill, size, shape, alpha, linetype * syntax: `scale_<aesthetic>_<type>` <img src="ggplot_presentation_assets/scales.png" alt="scales-syntax" style="width:50%;"> -- .pull-left-50[ ```r p <- ggplot(iris)+geom_point(aes(x=Sepal.Length, y=Sepal.Width,color=Species)) p ``` <img src="ggplot_presentation_files/figure-html/unnamed-chunk-30-1.svg" style="display: block; margin: auto auto auto 0;" /> ] -- .pull-right-50[ ```r p + scale_color_manual( name="Manual", values=c("#5BC0EB","#FDE74C","#9BC53D")) ``` <img src="ggplot_presentation_files/figure-html/unnamed-chunk-31-1.svg" style="display: block; margin: auto auto auto 0;" /> ] ??? Scales are used to control the aesthetics. For example the aesthetic color is mapped to a variable `x`. The palette of colors used, the mapping of which color to which value, the upper and lower limit of the data and colors etc is controlled by scales. --- name: scales-continuous-color ## Scales • Continuous Colors * In RStudio, type `scale_`, then press **TAB** -- .pull-left-50[ ```r p <- ggplot(iris)+ geom_point(aes(x=Sepal.Length, y=Sepal.Width, shape=Species,color=Petal.Length)) p ``` <img src="ggplot_presentation_files/figure-html/unnamed-chunk-32-1.svg" style="display: block; margin: auto auto auto 0;" /> ] -- .pull-right-50[ ```r p + scale_color_gradient(name="Pet Len", breaks=range(iris$Petal.Length), labels=c("Min","Max"), low="black",high="red") ``` <img src="ggplot_presentation_files/figure-html/unnamed-chunk-33-1.svg" style="display: block; margin: auto auto auto 0;" /> ] ??? Continuous colours can be changed using `scale_color_gradient()` for two colour gradient. Any number of breaks and colours can be specified using `scale_color_gradientn()`. --- name: scales-shape ## Scales • Shape .pull-left-50[ ```r p <- ggplot(iris)+ geom_point(aes(x=Sepal.Length, y=Sepal.Width, shape=Species,color=Species)) p ``` <img src="ggplot_presentation_files/figure-html/unnamed-chunk-34-1.svg" style="display: block; margin: auto auto auto 0;" /> ] -- .pull-right-50[ ```r p + scale_color_manual(name="New", values=c("blue","green","red"))+ scale_shape_manual(name="Bla",values=c(0,1,2)) ``` <img src="ggplot_presentation_files/figure-html/unnamed-chunk-35-1.svg" style="display: block; margin: auto auto auto 0;" /> ] ??? Shape scale can be adjusted using `scale_shape_manual()`. Multiple mappings for the same variable groups legends. --- name: scales-axis ## Scales • Axes * scales: x, y * syntax: `scale_<axis>_<type>` * arguments: name, limits, breaks, labels -- .pull-left-50[ ```r p <- ggplot(iris)+ geom_point(aes(x=Sepal.Length, y=Sepal.Width)) p ``` <img src="ggplot_presentation_files/figure-html/unnamed-chunk-36-1.svg" style="display: block; margin: auto auto auto 0;" /> ] -- .pull-right-50[ ```r p + scale_color_manual(name="New", values=c("blue","green","red"))+ scale_x_continuous(name="Sepal Length", breaks=seq(1,8),limits=c(3,5)) ``` <img src="ggplot_presentation_files/figure-html/unnamed-chunk-37-1.svg" style="display: block; margin: auto auto auto 0;" /> ] ??? The x and y axes are also controlled by scales. The axis break points, the break point text and limits are controlled through scales. When setting limits using `scale_`, the data outside the limits are dropped. Limits can also be set using `lims(x=c(3.5))` or `xlim(c(3,5))`. When mapping, `coord_map()` or `coord_cartesian()` is recommended for setting limits. --- name: facet-wrap ## Facets • `facet_wrap` * Split to subplots based on variable(s) * Facetting in one dimension -- .pull-left-50[ ```r p <- ggplot(iris)+ geom_point(aes(x=Sepal.Length, y=Sepal.Width, color=Species)) p ``` <img src="ggplot_presentation_files/figure-html/unnamed-chunk-38-1.svg" style="display: block; margin: auto auto auto 0;" /> ] -- .pull-right-50[ ```r p + facet_wrap(~Species) ``` <img src="ggplot_presentation_files/figure-html/unnamed-chunk-39-1.svg" style="display: block; margin: auto auto auto 0;" /> ```r p + facet_wrap(~Species,nrow=3) ``` <img src="ggplot_presentation_files/figure-html/unnamed-chunk-40-1.svg" style="display: block; margin: auto auto auto 0;" /> ] ??? `facet_wrap` is used to split a plot into subplots based on the categories in one or more variables. --- name: facet-grid ## Facets • `facet_grid` * Facetting in two dimensions .pull-left-50[ ```r p <- diamonds %>% ggplot(aes(carat,price))+ geom_point() p + facet_grid(~cut+clarity) ``` <img src="ggplot_presentation_files/figure-html/unnamed-chunk-41-1.svg" style="display: block; margin: auto auto auto 0;" /> ] -- .pull-left-50[ ```r p + facet_grid(cut~clarity) ``` <img src="ggplot_presentation_files/figure-html/unnamed-chunk-42-1.svg" style="display: block; margin: auto auto auto 0;" /> ] ??? `facet_grid` is also used to split a plot into subplots based on the categories in one or more variables. `facet_grid` can be used to create a matrix-like grid of two variables. --- name: coordinate ## Coordinate Systems 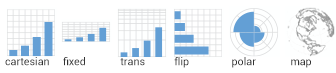 * `coord_cartesian(xlim=c(2,8))` for zooming in * `coord_map` for controlling limits on maps * `coord_polar` .pull-left-50[ ```r p <- ggplot(iris,aes(x="",y=Petal.Length,fill=Species))+ geom_bar(stat="identity") p ``` <img src="ggplot_presentation_files/figure-html/unnamed-chunk-43-1.svg" style="display: block; margin: auto auto auto 0;" /> ] ??? The coordinate system defines the surface used to represent numbers. Most plots use the cartesian coordinate sytem. Pie charts for example, is a polar coordinate projection of a cartesian barplot. Maps for example can have numerous coordinate systems called map projections. For example; UTM coordinates. -- .pull-right-50[ ```r p+coord_polar("y",start=0) ``` <img src="ggplot_presentation_files/figure-html/unnamed-chunk-44-1.svg" style="display: block; margin: auto auto auto 0;" /> ] --- name: theme ## Theme * Modify non-data plot elements/appearance * Axis labels, panel colors, legend appearance etc * Save a particular appearance for reuse * `?theme` -- .pull-left-50[ ```r ggplot(iris,aes(Petal.Length))+ geom_histogram()+ facet_wrap(~Species,nrow=2)+ theme_grey() ``` <img src="ggplot_presentation_files/figure-html/unnamed-chunk-45-1.svg" style="display: block; margin: auto auto auto 0;" /> ] -- .pull-right-50[ ```r ggplot(iris,aes(Petal.Length))+ geom_histogram()+ facet_wrap(~Species,nrow=2)+ theme_bw() ``` <img src="ggplot_presentation_files/figure-html/unnamed-chunk-46-1.svg" style="display: block; margin: auto auto auto 0;" /> ] ??? Themes allow to modify all non-data related components of the plot. This is the visual appearance of the plot. Examples include the axes line thickness, the background color or font family. --- name: theme-legend ## Theme • Legend ```r p <- ggplot(iris)+ geom_point(aes(x=Sepal.Length, y=Sepal.Width, color=Species)) ``` .pull-left-50[ ```r p + theme(legend.position="top") ``` <img src="ggplot_presentation_files/figure-html/unnamed-chunk-48-1.svg" style="display: block; margin: auto auto auto 0;" /> ] .pull-right-50[ ```r p + theme(legend.position="bottom") ``` <img src="ggplot_presentation_files/figure-html/unnamed-chunk-49-1.svg" style="display: block; margin: auto auto auto 0;" /> ] --- name: theme-text ## Theme • Text ```r element_text(family=NULL,face=NULL,color=NULL,size=NULL,hjust=NULL, vjust=NULL, angle=NULL,lineheight=NULL,margin = NULL) ``` ```r p <- p + theme( axis.title=element_text(color="#e41a1c"), axis.text=element_text(color="#377eb8"), plot.title=element_text(color="#4daf4a"), plot.subtitle=element_text(color="#984ea3"), legend.text=element_text(color="#ff7f00"), legend.title=element_text(color="#ffff33"), strip.text=element_text(color="#a65628") ) ``` <img src="ggplot_presentation_files/figure-html/unnamed-chunk-53-1.svg" style="display: block; margin: auto auto auto 0;" /> --- name: theme-rect ## Theme • Rect ```r element_rect(fill=NULL,color=NULL,size=NULL,linetype=NULL) ``` ```r p <- p + theme( plot.background=element_rect(fill="#b3e2cd"), panel.background=element_rect(fill="#fdcdac"), panel.border=element_rect(fill=NA,color="#cbd5e8",size=3), legend.background=element_rect(fill="#f4cae4"), legend.box.background=element_rect(fill="#e6f5c9"), strip.background=element_rect(fill="#fff2ae") ) ``` <img src="ggplot_presentation_files/figure-html/unnamed-chunk-57-1.svg" style="display: block; margin: auto auto auto 0;" /> --- name: theme-save ## Theme • Reuse ```r newtheme <- theme_bw() + theme( axis.ticks=element_blank(), panel.background=element_rect(fill="white"), panel.grid.minor=element_blank(), panel.grid.major.x=element_blank(), panel.grid.major.y=element_line(size=0.3,color="grey90"), panel.border=element_blank(), legend.position="top", legend.justification="right" ) ``` .pull-left-50[ ```r p ``` <img src="ggplot_presentation_files/figure-html/unnamed-chunk-60-1.svg" style="display: block; margin: auto auto auto 0;" /> ] .pull-right-50[ ```r p + newtheme ``` <img src="ggplot_presentation_files/figure-html/unnamed-chunk-61-1.svg" style="display: block; margin: auto auto auto 0;" /> ] --- name: position ## Position <table class="table table-striped table-hover table-responsive table-condensed" style="width: auto !important; "> <thead> <tr> <th style="text-align:left;"> </th> <th style="text-align:center;"> Murder </th> <th style="text-align:center;"> Assault </th> <th style="text-align:center;"> UrbanPop </th> <th style="text-align:center;"> Rape </th> </tr> </thead> <tbody> <tr> <td style="text-align:left;"> Alabama </td> <td style="text-align:center;"> 13.2 </td> <td style="text-align:center;"> 236 </td> <td style="text-align:center;"> 58 </td> <td style="text-align:center;"> 21.2 </td> </tr> <tr> <td style="text-align:left;"> Alaska </td> <td style="text-align:center;"> 10.0 </td> <td style="text-align:center;"> 263 </td> <td style="text-align:center;"> 48 </td> <td style="text-align:center;"> 44.5 </td> </tr> <tr> <td style="text-align:left;"> Arizona </td> <td style="text-align:center;"> 8.1 </td> <td style="text-align:center;"> 294 </td> <td style="text-align:center;"> 80 </td> <td style="text-align:center;"> 31.0 </td> </tr> </tbody> </table> ```r us <- USArrests %>% mutate(state=rownames(.)) %>% slice(1:4) %>% gather(key=type,value=value,-state) p <- ggplot(us,aes(x=state,y=value,fill=type)) ``` -- .pull-left-50[ ```r p + geom_bar(stat="identity",position="stack") ``` <img src="ggplot_presentation_files/figure-html/unnamed-chunk-64-1.svg" style="display: block; margin: auto auto auto 0;" /> ] -- .pull-right-50[ ```r p + geom_bar(stat="identity",position="dodge") ``` <img src="ggplot_presentation_files/figure-html/unnamed-chunk-65-1.svg" style="display: block; margin: auto auto auto 0;" /> ] --- name: save ## Saving plots ```r p <- ggplot(iris,aes(Petal.Length,Sepal.Length,color=Species))+ geom_point() ``` * `ggplot2` plots can be saved just like base plots ```r png("plot.png",height=5,width=7,units="cm",res=200) print(p) dev.off() ``` * `ggplot2` package offers a convenient function ```r ggsave("plot.png",p,height=5,width=7,units="cm",dpi=200,type="cairo") ``` * Use `type="cairo"` for nicer anti-aliasing * Note that default units in `png` is pixels while in `ggsave` it's inches --- name: comb ## Combining Plots ```r p <- ggplot(us,aes(x=state,y=value,color=type))+geom_point() q <- ggplot(us,aes(x=state,y=value,fill=type))+geom_bar(stat="identity") ``` ```r gridExtra::grid.arrange(p,q,ncol=2) ``` <img src="ggplot_presentation_files/figure-html/unnamed-chunk-71-1.svg" style="display: block; margin: auto auto auto 0;" /> -- -- ```r cowplot::plot_grid() ``` ??? Combining two or more `ggplot2` plots is often required and several packages exist to help with this situation. Some functions allow plots to be placed adjacently, also allowing varying heights or widths of each plot. Some functions allow one plot to be plotted on another plot like a subset plot. --- name: interactive ## Interactive * Convert `ggplot2` object to interactive HTML ```r p <- ggplot(iris,aes(x=Sepal.Length,y=Sepal.Width,col=Species)) ``` .pull-left-50[ ```r p1 <- p+geom_point() plotly::ggplotly(p1,width=400,height=300) ``` <div id="htmlwidget-b3ed89cc5bcffd47fcf1" style="width:400px;height:300px;" class="plotly html-widget"></div> <script type="application/json" data-for="htmlwidget-b3ed89cc5bcffd47fcf1">{"x":{"data":[{"x":[5.1,4.9,4.7,4.6,5,5.4,4.6,5,4.4,4.9,5.4,4.8,4.8,4.3,5.8,5.7,5.4,5.1,5.7,5.1,5.4,5.1,4.6,5.1,4.8,5,5,5.2,5.2,4.7,4.8,5.4,5.2,5.5,4.9,5,5.5,4.9,4.4,5.1,5,4.5,4.4,5,5.1,4.8,5.1,4.6,5.3,5],"y":[3.5,3,3.2,3.1,3.6,3.9,3.4,3.4,2.9,3.1,3.7,3.4,3,3,4,4.4,3.9,3.5,3.8,3.8,3.4,3.7,3.6,3.3,3.4,3,3.4,3.5,3.4,3.2,3.1,3.4,4.1,4.2,3.1,3.2,3.5,3.6,3,3.4,3.5,2.3,3.2,3.5,3.8,3,3.8,3.2,3.7,3.3],"text":["Sepal.Length: 5.1<br />Sepal.Width: 3.5<br />Species: setosa","Sepal.Length: 4.9<br />Sepal.Width: 3.0<br />Species: setosa","Sepal.Length: 4.7<br />Sepal.Width: 3.2<br />Species: setosa","Sepal.Length: 4.6<br />Sepal.Width: 3.1<br />Species: setosa","Sepal.Length: 5.0<br />Sepal.Width: 3.6<br />Species: setosa","Sepal.Length: 5.4<br />Sepal.Width: 3.9<br />Species: setosa","Sepal.Length: 4.6<br />Sepal.Width: 3.4<br />Species: setosa","Sepal.Length: 5.0<br />Sepal.Width: 3.4<br />Species: setosa","Sepal.Length: 4.4<br />Sepal.Width: 2.9<br />Species: setosa","Sepal.Length: 4.9<br />Sepal.Width: 3.1<br />Species: setosa","Sepal.Length: 5.4<br />Sepal.Width: 3.7<br />Species: setosa","Sepal.Length: 4.8<br />Sepal.Width: 3.4<br />Species: setosa","Sepal.Length: 4.8<br />Sepal.Width: 3.0<br />Species: setosa","Sepal.Length: 4.3<br />Sepal.Width: 3.0<br />Species: setosa","Sepal.Length: 5.8<br />Sepal.Width: 4.0<br />Species: setosa","Sepal.Length: 5.7<br />Sepal.Width: 4.4<br />Species: setosa","Sepal.Length: 5.4<br />Sepal.Width: 3.9<br />Species: setosa","Sepal.Length: 5.1<br />Sepal.Width: 3.5<br />Species: setosa","Sepal.Length: 5.7<br />Sepal.Width: 3.8<br />Species: setosa","Sepal.Length: 5.1<br />Sepal.Width: 3.8<br />Species: setosa","Sepal.Length: 5.4<br />Sepal.Width: 3.4<br />Species: setosa","Sepal.Length: 5.1<br />Sepal.Width: 3.7<br />Species: setosa","Sepal.Length: 4.6<br />Sepal.Width: 3.6<br />Species: setosa","Sepal.Length: 5.1<br />Sepal.Width: 3.3<br />Species: setosa","Sepal.Length: 4.8<br />Sepal.Width: 3.4<br />Species: setosa","Sepal.Length: 5.0<br />Sepal.Width: 3.0<br />Species: setosa","Sepal.Length: 5.0<br />Sepal.Width: 3.4<br />Species: setosa","Sepal.Length: 5.2<br />Sepal.Width: 3.5<br />Species: setosa","Sepal.Length: 5.2<br />Sepal.Width: 3.4<br />Species: setosa","Sepal.Length: 4.7<br />Sepal.Width: 3.2<br />Species: setosa","Sepal.Length: 4.8<br />Sepal.Width: 3.1<br />Species: setosa","Sepal.Length: 5.4<br />Sepal.Width: 3.4<br />Species: setosa","Sepal.Length: 5.2<br />Sepal.Width: 4.1<br />Species: setosa","Sepal.Length: 5.5<br />Sepal.Width: 4.2<br />Species: setosa","Sepal.Length: 4.9<br />Sepal.Width: 3.1<br />Species: setosa","Sepal.Length: 5.0<br />Sepal.Width: 3.2<br />Species: setosa","Sepal.Length: 5.5<br />Sepal.Width: 3.5<br />Species: setosa","Sepal.Length: 4.9<br />Sepal.Width: 3.6<br />Species: setosa","Sepal.Length: 4.4<br />Sepal.Width: 3.0<br />Species: setosa","Sepal.Length: 5.1<br />Sepal.Width: 3.4<br />Species: setosa","Sepal.Length: 5.0<br />Sepal.Width: 3.5<br />Species: setosa","Sepal.Length: 4.5<br />Sepal.Width: 2.3<br />Species: setosa","Sepal.Length: 4.4<br />Sepal.Width: 3.2<br />Species: setosa","Sepal.Length: 5.0<br />Sepal.Width: 3.5<br />Species: setosa","Sepal.Length: 5.1<br />Sepal.Width: 3.8<br />Species: setosa","Sepal.Length: 4.8<br />Sepal.Width: 3.0<br />Species: setosa","Sepal.Length: 5.1<br />Sepal.Width: 3.8<br />Species: setosa","Sepal.Length: 4.6<br />Sepal.Width: 3.2<br />Species: setosa","Sepal.Length: 5.3<br />Sepal.Width: 3.7<br />Species: setosa","Sepal.Length: 5.0<br />Sepal.Width: 3.3<br />Species: setosa"],"type":"scatter","mode":"markers","marker":{"autocolorscale":false,"color":"rgba(248,118,109,1)","opacity":1,"size":5.66929133858268,"symbol":"circle","line":{"width":1.88976377952756,"color":"rgba(248,118,109,1)"}},"hoveron":"points","name":"setosa","legendgroup":"setosa","showlegend":true,"xaxis":"x","yaxis":"y","hoverinfo":"text","frame":null},{"x":[7,6.4,6.9,5.5,6.5,5.7,6.3,4.9,6.6,5.2,5,5.9,6,6.1,5.6,6.7,5.6,5.8,6.2,5.6,5.9,6.1,6.3,6.1,6.4,6.6,6.8,6.7,6,5.7,5.5,5.5,5.8,6,5.4,6,6.7,6.3,5.6,5.5,5.5,6.1,5.8,5,5.6,5.7,5.7,6.2,5.1,5.7],"y":[3.2,3.2,3.1,2.3,2.8,2.8,3.3,2.4,2.9,2.7,2,3,2.2,2.9,2.9,3.1,3,2.7,2.2,2.5,3.2,2.8,2.5,2.8,2.9,3,2.8,3,2.9,2.6,2.4,2.4,2.7,2.7,3,3.4,3.1,2.3,3,2.5,2.6,3,2.6,2.3,2.7,3,2.9,2.9,2.5,2.8],"text":["Sepal.Length: 7.0<br />Sepal.Width: 3.2<br />Species: versicolor","Sepal.Length: 6.4<br />Sepal.Width: 3.2<br />Species: versicolor","Sepal.Length: 6.9<br />Sepal.Width: 3.1<br />Species: versicolor","Sepal.Length: 5.5<br />Sepal.Width: 2.3<br />Species: versicolor","Sepal.Length: 6.5<br />Sepal.Width: 2.8<br />Species: versicolor","Sepal.Length: 5.7<br />Sepal.Width: 2.8<br />Species: versicolor","Sepal.Length: 6.3<br />Sepal.Width: 3.3<br />Species: versicolor","Sepal.Length: 4.9<br />Sepal.Width: 2.4<br />Species: versicolor","Sepal.Length: 6.6<br />Sepal.Width: 2.9<br />Species: versicolor","Sepal.Length: 5.2<br />Sepal.Width: 2.7<br />Species: versicolor","Sepal.Length: 5.0<br />Sepal.Width: 2.0<br />Species: versicolor","Sepal.Length: 5.9<br />Sepal.Width: 3.0<br />Species: versicolor","Sepal.Length: 6.0<br />Sepal.Width: 2.2<br />Species: versicolor","Sepal.Length: 6.1<br />Sepal.Width: 2.9<br />Species: versicolor","Sepal.Length: 5.6<br />Sepal.Width: 2.9<br />Species: versicolor","Sepal.Length: 6.7<br />Sepal.Width: 3.1<br />Species: versicolor","Sepal.Length: 5.6<br />Sepal.Width: 3.0<br />Species: versicolor","Sepal.Length: 5.8<br />Sepal.Width: 2.7<br />Species: versicolor","Sepal.Length: 6.2<br />Sepal.Width: 2.2<br />Species: versicolor","Sepal.Length: 5.6<br />Sepal.Width: 2.5<br />Species: versicolor","Sepal.Length: 5.9<br />Sepal.Width: 3.2<br />Species: versicolor","Sepal.Length: 6.1<br />Sepal.Width: 2.8<br />Species: versicolor","Sepal.Length: 6.3<br />Sepal.Width: 2.5<br />Species: versicolor","Sepal.Length: 6.1<br />Sepal.Width: 2.8<br />Species: versicolor","Sepal.Length: 6.4<br />Sepal.Width: 2.9<br />Species: versicolor","Sepal.Length: 6.6<br />Sepal.Width: 3.0<br />Species: versicolor","Sepal.Length: 6.8<br />Sepal.Width: 2.8<br />Species: versicolor","Sepal.Length: 6.7<br />Sepal.Width: 3.0<br />Species: versicolor","Sepal.Length: 6.0<br />Sepal.Width: 2.9<br />Species: versicolor","Sepal.Length: 5.7<br />Sepal.Width: 2.6<br />Species: versicolor","Sepal.Length: 5.5<br />Sepal.Width: 2.4<br />Species: versicolor","Sepal.Length: 5.5<br />Sepal.Width: 2.4<br />Species: versicolor","Sepal.Length: 5.8<br />Sepal.Width: 2.7<br />Species: versicolor","Sepal.Length: 6.0<br />Sepal.Width: 2.7<br />Species: versicolor","Sepal.Length: 5.4<br />Sepal.Width: 3.0<br />Species: versicolor","Sepal.Length: 6.0<br />Sepal.Width: 3.4<br />Species: versicolor","Sepal.Length: 6.7<br />Sepal.Width: 3.1<br />Species: versicolor","Sepal.Length: 6.3<br />Sepal.Width: 2.3<br />Species: versicolor","Sepal.Length: 5.6<br />Sepal.Width: 3.0<br />Species: versicolor","Sepal.Length: 5.5<br />Sepal.Width: 2.5<br />Species: versicolor","Sepal.Length: 5.5<br />Sepal.Width: 2.6<br />Species: versicolor","Sepal.Length: 6.1<br />Sepal.Width: 3.0<br />Species: versicolor","Sepal.Length: 5.8<br />Sepal.Width: 2.6<br />Species: versicolor","Sepal.Length: 5.0<br />Sepal.Width: 2.3<br />Species: versicolor","Sepal.Length: 5.6<br />Sepal.Width: 2.7<br />Species: versicolor","Sepal.Length: 5.7<br />Sepal.Width: 3.0<br />Species: versicolor","Sepal.Length: 5.7<br />Sepal.Width: 2.9<br />Species: versicolor","Sepal.Length: 6.2<br />Sepal.Width: 2.9<br />Species: versicolor","Sepal.Length: 5.1<br />Sepal.Width: 2.5<br />Species: versicolor","Sepal.Length: 5.7<br />Sepal.Width: 2.8<br />Species: versicolor"],"type":"scatter","mode":"markers","marker":{"autocolorscale":false,"color":"rgba(0,186,56,1)","opacity":1,"size":5.66929133858268,"symbol":"circle","line":{"width":1.88976377952756,"color":"rgba(0,186,56,1)"}},"hoveron":"points","name":"versicolor","legendgroup":"versicolor","showlegend":true,"xaxis":"x","yaxis":"y","hoverinfo":"text","frame":null},{"x":[6.3,5.8,7.1,6.3,6.5,7.6,4.9,7.3,6.7,7.2,6.5,6.4,6.8,5.7,5.8,6.4,6.5,7.7,7.7,6,6.9,5.6,7.7,6.3,6.7,7.2,6.2,6.1,6.4,7.2,7.4,7.9,6.4,6.3,6.1,7.7,6.3,6.4,6,6.9,6.7,6.9,5.8,6.8,6.7,6.7,6.3,6.5,6.2,5.9],"y":[3.3,2.7,3,2.9,3,3,2.5,2.9,2.5,3.6,3.2,2.7,3,2.5,2.8,3.2,3,3.8,2.6,2.2,3.2,2.8,2.8,2.7,3.3,3.2,2.8,3,2.8,3,2.8,3.8,2.8,2.8,2.6,3,3.4,3.1,3,3.1,3.1,3.1,2.7,3.2,3.3,3,2.5,3,3.4,3],"text":["Sepal.Length: 6.3<br />Sepal.Width: 3.3<br />Species: virginica","Sepal.Length: 5.8<br />Sepal.Width: 2.7<br />Species: virginica","Sepal.Length: 7.1<br />Sepal.Width: 3.0<br />Species: virginica","Sepal.Length: 6.3<br />Sepal.Width: 2.9<br />Species: virginica","Sepal.Length: 6.5<br />Sepal.Width: 3.0<br />Species: virginica","Sepal.Length: 7.6<br />Sepal.Width: 3.0<br />Species: virginica","Sepal.Length: 4.9<br />Sepal.Width: 2.5<br />Species: virginica","Sepal.Length: 7.3<br />Sepal.Width: 2.9<br />Species: virginica","Sepal.Length: 6.7<br />Sepal.Width: 2.5<br />Species: virginica","Sepal.Length: 7.2<br />Sepal.Width: 3.6<br />Species: virginica","Sepal.Length: 6.5<br />Sepal.Width: 3.2<br />Species: virginica","Sepal.Length: 6.4<br />Sepal.Width: 2.7<br />Species: virginica","Sepal.Length: 6.8<br />Sepal.Width: 3.0<br />Species: virginica","Sepal.Length: 5.7<br />Sepal.Width: 2.5<br />Species: virginica","Sepal.Length: 5.8<br />Sepal.Width: 2.8<br />Species: virginica","Sepal.Length: 6.4<br />Sepal.Width: 3.2<br />Species: virginica","Sepal.Length: 6.5<br />Sepal.Width: 3.0<br />Species: virginica","Sepal.Length: 7.7<br />Sepal.Width: 3.8<br />Species: virginica","Sepal.Length: 7.7<br />Sepal.Width: 2.6<br />Species: virginica","Sepal.Length: 6.0<br />Sepal.Width: 2.2<br />Species: virginica","Sepal.Length: 6.9<br />Sepal.Width: 3.2<br />Species: virginica","Sepal.Length: 5.6<br />Sepal.Width: 2.8<br />Species: virginica","Sepal.Length: 7.7<br />Sepal.Width: 2.8<br />Species: virginica","Sepal.Length: 6.3<br />Sepal.Width: 2.7<br />Species: virginica","Sepal.Length: 6.7<br />Sepal.Width: 3.3<br />Species: virginica","Sepal.Length: 7.2<br />Sepal.Width: 3.2<br />Species: virginica","Sepal.Length: 6.2<br />Sepal.Width: 2.8<br />Species: virginica","Sepal.Length: 6.1<br />Sepal.Width: 3.0<br />Species: virginica","Sepal.Length: 6.4<br />Sepal.Width: 2.8<br />Species: virginica","Sepal.Length: 7.2<br />Sepal.Width: 3.0<br />Species: virginica","Sepal.Length: 7.4<br />Sepal.Width: 2.8<br />Species: virginica","Sepal.Length: 7.9<br />Sepal.Width: 3.8<br />Species: virginica","Sepal.Length: 6.4<br />Sepal.Width: 2.8<br />Species: virginica","Sepal.Length: 6.3<br />Sepal.Width: 2.8<br />Species: virginica","Sepal.Length: 6.1<br />Sepal.Width: 2.6<br />Species: virginica","Sepal.Length: 7.7<br />Sepal.Width: 3.0<br />Species: virginica","Sepal.Length: 6.3<br />Sepal.Width: 3.4<br />Species: virginica","Sepal.Length: 6.4<br />Sepal.Width: 3.1<br />Species: virginica","Sepal.Length: 6.0<br />Sepal.Width: 3.0<br />Species: virginica","Sepal.Length: 6.9<br />Sepal.Width: 3.1<br />Species: virginica","Sepal.Length: 6.7<br />Sepal.Width: 3.1<br />Species: virginica","Sepal.Length: 6.9<br />Sepal.Width: 3.1<br />Species: virginica","Sepal.Length: 5.8<br />Sepal.Width: 2.7<br />Species: virginica","Sepal.Length: 6.8<br />Sepal.Width: 3.2<br />Species: virginica","Sepal.Length: 6.7<br />Sepal.Width: 3.3<br />Species: virginica","Sepal.Length: 6.7<br />Sepal.Width: 3.0<br />Species: virginica","Sepal.Length: 6.3<br />Sepal.Width: 2.5<br />Species: virginica","Sepal.Length: 6.5<br />Sepal.Width: 3.0<br />Species: virginica","Sepal.Length: 6.2<br />Sepal.Width: 3.4<br />Species: virginica","Sepal.Length: 5.9<br />Sepal.Width: 3.0<br />Species: virginica"],"type":"scatter","mode":"markers","marker":{"autocolorscale":false,"color":"rgba(97,156,255,1)","opacity":1,"size":5.66929133858268,"symbol":"circle","line":{"width":1.88976377952756,"color":"rgba(97,156,255,1)"}},"hoveron":"points","name":"virginica","legendgroup":"virginica","showlegend":true,"xaxis":"x","yaxis":"y","hoverinfo":"text","frame":null}],"layout":{"margin":{"t":25.7412480974125,"r":7.30593607305936,"b":39.6955859969559,"l":43.1050228310502},"plot_bgcolor":"rgba(235,235,235,1)","paper_bgcolor":"rgba(255,255,255,1)","font":{"color":"rgba(0,0,0,1)","family":"","size":14.6118721461187},"xaxis":{"domain":[0,1],"automargin":true,"type":"linear","autorange":false,"range":[4.12,8.08],"tickmode":"array","ticktext":["5","6","7","8"],"tickvals":[5,6,7,8],"categoryorder":"array","categoryarray":["5","6","7","8"],"nticks":null,"ticks":"outside","tickcolor":"rgba(51,51,51,1)","ticklen":3.65296803652968,"tickwidth":0.66417600664176,"showticklabels":true,"tickfont":{"color":"rgba(77,77,77,1)","family":"","size":11.689497716895},"tickangle":-0,"showline":false,"linecolor":null,"linewidth":0,"showgrid":true,"gridcolor":"rgba(255,255,255,1)","gridwidth":0.66417600664176,"zeroline":false,"anchor":"y","title":{"text":"Sepal.Length","font":{"color":"rgba(0,0,0,1)","family":"","size":14.6118721461187}},"hoverformat":".2f"},"yaxis":{"domain":[0,1],"automargin":true,"type":"linear","autorange":false,"range":[1.88,4.52],"tickmode":"array","ticktext":["2.0","2.5","3.0","3.5","4.0","4.5"],"tickvals":[2,2.5,3,3.5,4,4.5],"categoryorder":"array","categoryarray":["2.0","2.5","3.0","3.5","4.0","4.5"],"nticks":null,"ticks":"outside","tickcolor":"rgba(51,51,51,1)","ticklen":3.65296803652968,"tickwidth":0.66417600664176,"showticklabels":true,"tickfont":{"color":"rgba(77,77,77,1)","family":"","size":11.689497716895},"tickangle":-0,"showline":false,"linecolor":null,"linewidth":0,"showgrid":true,"gridcolor":"rgba(255,255,255,1)","gridwidth":0.66417600664176,"zeroline":false,"anchor":"x","title":{"text":"Sepal.Width","font":{"color":"rgba(0,0,0,1)","family":"","size":14.6118721461187}},"hoverformat":".2f"},"shapes":[{"type":"rect","fillcolor":null,"line":{"color":null,"width":0,"linetype":[]},"yref":"paper","xref":"paper","x0":0,"x1":1,"y0":0,"y1":1}],"showlegend":true,"legend":{"bgcolor":"rgba(255,255,255,1)","bordercolor":"transparent","borderwidth":1.88976377952756,"font":{"color":"rgba(0,0,0,1)","family":"","size":11.689497716895},"y":0.896062992125984},"annotations":[{"text":"Species","x":1.02,"y":1,"showarrow":false,"ax":0,"ay":0,"font":{"color":"rgba(0,0,0,1)","family":"","size":14.6118721461187},"xref":"paper","yref":"paper","textangle":-0,"xanchor":"left","yanchor":"bottom","legendTitle":true}],"hovermode":"closest","width":400,"height":300,"barmode":"relative"},"config":{"doubleClick":"reset","showSendToCloud":false},"source":"A","attrs":{"e299cbf90e1":{"x":{},"y":{},"colour":{},"type":"scatter"}},"cur_data":"e299cbf90e1","visdat":{"e299cbf90e1":["function (y) ","x"]},"highlight":{"on":"plotly_click","persistent":false,"dynamic":false,"selectize":false,"opacityDim":0.2,"selected":{"opacity":1},"debounce":0},"shinyEvents":["plotly_hover","plotly_click","plotly_selected","plotly_relayout","plotly_brushed","plotly_brushing","plotly_clickannotation","plotly_doubleclick","plotly_deselect","plotly_afterplot","plotly_sunburstclick"],"base_url":"https://plot.ly"},"evals":[],"jsHooks":[]}</script> ] .pull-right-50[ ```r p2 <- p+ggiraph::geom_point_interactive( aes(tooltip=paste0("<b>Species: </b>",Species)), size=2)+theme_bw(base_size=12) ggiraph::ggiraph(code=print(p2)) ``` <div id="htmlwidget-ac108ed8f735ae5db717" style="width:324px;height:324px;" class="girafe html-widget"></div> <script type="application/json" data-for="htmlwidget-ac108ed8f735ae5db717">{"x":{"html":"<?xml version=\"1.0\" encoding=\"UTF-8\"?>\n<svg xmlns='http://www.w3.org/2000/svg' xmlns:xlink='http://www.w3.org/1999/xlink' id='svg_f97619c2-9532-4b85-acc2-030cfa5af1d9' viewBox='0 0 432.00 360.00'>\n <g>\n <defs>\n <clipPath id='svg_f97619c2-9532-4b85-acc2-030cfa5af1d9_cl_1'>\n <rect x='0.00' y='0.00' width='432.00' height='360.00'/>\n <\/clipPath>\n <\/defs>\n <rect x='0.00' y='0.00' width='432.00' height='360.00' id='svg_f97619c2-9532-4b85-acc2-030cfa5af1d9_el_1' clip-path='url(#svg_f97619c2-9532-4b85-acc2-030cfa5af1d9_cl_1)' fill='#FFFFFF' fill-opacity='1' stroke='#FFFFFF' stroke-opacity='1' stroke-width='0.75' stroke-linejoin='round' stroke-linecap='round'/>\n <defs>\n <clipPath id='svg_f97619c2-9532-4b85-acc2-030cfa5af1d9_cl_2'>\n <rect x='0.00' y='0.00' width='432.00' height='360.00'/>\n <\/clipPath>\n <\/defs>\n <rect x='0.00' y='0.00' width='432.00' height='360.00' id='svg_f97619c2-9532-4b85-acc2-030cfa5af1d9_el_2' clip-path='url(#svg_f97619c2-9532-4b85-acc2-030cfa5af1d9_cl_2)' fill='#FFFFFF' fill-opacity='1' stroke='#FFFFFF' stroke-opacity='1' stroke-width='1.16' stroke-linejoin='round' stroke-linecap='round'/>\n <defs>\n <clipPath id='svg_f97619c2-9532-4b85-acc2-030cfa5af1d9_cl_3'>\n <rect x='40.84' y='5.98' width='291.22' height='319.44'/>\n <\/clipPath>\n <\/defs>\n <rect x='40.84' y='5.98' width='291.22' height='319.44' id='svg_f97619c2-9532-4b85-acc2-030cfa5af1d9_el_3' clip-path='url(#svg_f97619c2-9532-4b85-acc2-030cfa5af1d9_cl_3)' fill='#FFFFFF' fill-opacity='1' stroke='none'/>\n <polyline points='40.84,280.65 332.05,280.65' id='svg_f97619c2-9532-4b85-acc2-030cfa5af1d9_el_4' clip-path='url(#svg_f97619c2-9532-4b85-acc2-030cfa5af1d9_cl_3)' fill='none' stroke='#EBEBEB' stroke-opacity='1' stroke-width='0.58' stroke-linejoin='round' stroke-linecap='butt'/>\n <polyline points='40.84,220.15 332.05,220.15' id='svg_f97619c2-9532-4b85-acc2-030cfa5af1d9_el_5' clip-path='url(#svg_f97619c2-9532-4b85-acc2-030cfa5af1d9_cl_3)' fill='none' stroke='#EBEBEB' stroke-opacity='1' stroke-width='0.58' stroke-linejoin='round' stroke-linecap='butt'/>\n <polyline points='40.84,159.65 332.05,159.65' id='svg_f97619c2-9532-4b85-acc2-030cfa5af1d9_el_6' clip-path='url(#svg_f97619c2-9532-4b85-acc2-030cfa5af1d9_cl_3)' fill='none' stroke='#EBEBEB' stroke-opacity='1' stroke-width='0.58' stroke-linejoin='round' stroke-linecap='butt'/>\n <polyline points='40.84,99.15 332.05,99.15' id='svg_f97619c2-9532-4b85-acc2-030cfa5af1d9_el_7' clip-path='url(#svg_f97619c2-9532-4b85-acc2-030cfa5af1d9_cl_3)' fill='none' stroke='#EBEBEB' stroke-opacity='1' stroke-width='0.58' stroke-linejoin='round' stroke-linecap='butt'/>\n <polyline points='40.84,38.65 332.05,38.65' id='svg_f97619c2-9532-4b85-acc2-030cfa5af1d9_el_8' clip-path='url(#svg_f97619c2-9532-4b85-acc2-030cfa5af1d9_cl_3)' fill='none' stroke='#EBEBEB' stroke-opacity='1' stroke-width='0.58' stroke-linejoin='round' stroke-linecap='butt'/>\n <polyline points='68.78,325.42 68.78,5.98' id='svg_f97619c2-9532-4b85-acc2-030cfa5af1d9_el_9' clip-path='url(#svg_f97619c2-9532-4b85-acc2-030cfa5af1d9_cl_3)' fill='none' stroke='#EBEBEB' stroke-opacity='1' stroke-width='0.58' stroke-linejoin='round' stroke-linecap='butt'/>\n <polyline points='142.32,325.42 142.32,5.98' id='svg_f97619c2-9532-4b85-acc2-030cfa5af1d9_el_10' clip-path='url(#svg_f97619c2-9532-4b85-acc2-030cfa5af1d9_cl_3)' fill='none' stroke='#EBEBEB' stroke-opacity='1' stroke-width='0.58' stroke-linejoin='round' stroke-linecap='butt'/>\n <polyline points='215.86,325.42 215.86,5.98' id='svg_f97619c2-9532-4b85-acc2-030cfa5af1d9_el_11' clip-path='url(#svg_f97619c2-9532-4b85-acc2-030cfa5af1d9_cl_3)' fill='none' stroke='#EBEBEB' stroke-opacity='1' stroke-width='0.58' stroke-linejoin='round' stroke-linecap='butt'/>\n <polyline points='289.40,325.42 289.40,5.98' id='svg_f97619c2-9532-4b85-acc2-030cfa5af1d9_el_12' clip-path='url(#svg_f97619c2-9532-4b85-acc2-030cfa5af1d9_cl_3)' fill='none' stroke='#EBEBEB' stroke-opacity='1' stroke-width='0.58' stroke-linejoin='round' stroke-linecap='butt'/>\n <polyline points='40.84,310.90 332.05,310.90' id='svg_f97619c2-9532-4b85-acc2-030cfa5af1d9_el_13' clip-path='url(#svg_f97619c2-9532-4b85-acc2-030cfa5af1d9_cl_3)' fill='none' stroke='#EBEBEB' stroke-opacity='1' stroke-width='1.16' stroke-linejoin='round' stroke-linecap='butt'/>\n <polyline points='40.84,250.40 332.05,250.40' id='svg_f97619c2-9532-4b85-acc2-030cfa5af1d9_el_14' clip-path='url(#svg_f97619c2-9532-4b85-acc2-030cfa5af1d9_cl_3)' fill='none' stroke='#EBEBEB' stroke-opacity='1' stroke-width='1.16' stroke-linejoin='round' stroke-linecap='butt'/>\n <polyline points='40.84,189.90 332.05,189.90' id='svg_f97619c2-9532-4b85-acc2-030cfa5af1d9_el_15' clip-path='url(#svg_f97619c2-9532-4b85-acc2-030cfa5af1d9_cl_3)' fill='none' stroke='#EBEBEB' stroke-opacity='1' stroke-width='1.16' stroke-linejoin='round' stroke-linecap='butt'/>\n <polyline points='40.84,129.40 332.05,129.40' id='svg_f97619c2-9532-4b85-acc2-030cfa5af1d9_el_16' clip-path='url(#svg_f97619c2-9532-4b85-acc2-030cfa5af1d9_cl_3)' fill='none' stroke='#EBEBEB' stroke-opacity='1' stroke-width='1.16' stroke-linejoin='round' stroke-linecap='butt'/>\n <polyline points='40.84,68.90 332.05,68.90' id='svg_f97619c2-9532-4b85-acc2-030cfa5af1d9_el_17' clip-path='url(#svg_f97619c2-9532-4b85-acc2-030cfa5af1d9_cl_3)' fill='none' stroke='#EBEBEB' stroke-opacity='1' stroke-width='1.16' stroke-linejoin='round' stroke-linecap='butt'/>\n <polyline points='40.84,8.40 332.05,8.40' id='svg_f97619c2-9532-4b85-acc2-030cfa5af1d9_el_18' clip-path='url(#svg_f97619c2-9532-4b85-acc2-030cfa5af1d9_cl_3)' fill='none' stroke='#EBEBEB' stroke-opacity='1' stroke-width='1.16' stroke-linejoin='round' stroke-linecap='butt'/>\n <polyline points='105.55,325.42 105.55,5.98' id='svg_f97619c2-9532-4b85-acc2-030cfa5af1d9_el_19' clip-path='url(#svg_f97619c2-9532-4b85-acc2-030cfa5af1d9_cl_3)' fill='none' stroke='#EBEBEB' stroke-opacity='1' stroke-width='1.16' stroke-linejoin='round' stroke-linecap='butt'/>\n <polyline points='179.09,325.42 179.09,5.98' id='svg_f97619c2-9532-4b85-acc2-030cfa5af1d9_el_20' clip-path='url(#svg_f97619c2-9532-4b85-acc2-030cfa5af1d9_cl_3)' fill='none' stroke='#EBEBEB' stroke-opacity='1' stroke-width='1.16' stroke-linejoin='round' stroke-linecap='butt'/>\n <polyline points='252.63,325.42 252.63,5.98' id='svg_f97619c2-9532-4b85-acc2-030cfa5af1d9_el_21' clip-path='url(#svg_f97619c2-9532-4b85-acc2-030cfa5af1d9_cl_3)' fill='none' stroke='#EBEBEB' stroke-opacity='1' stroke-width='1.16' stroke-linejoin='round' stroke-linecap='butt'/>\n <polyline points='326.17,325.42 326.17,5.98' id='svg_f97619c2-9532-4b85-acc2-030cfa5af1d9_el_22' clip-path='url(#svg_f97619c2-9532-4b85-acc2-030cfa5af1d9_cl_3)' fill='none' stroke='#EBEBEB' stroke-opacity='1' stroke-width='1.16' stroke-linejoin='round' stroke-linecap='butt'/>\n <circle cx='112.90' cy='129.40' r='1.87pt' id='svg_f97619c2-9532-4b85-acc2-030cfa5af1d9_el_23' clip-path='url(#svg_f97619c2-9532-4b85-acc2-030cfa5af1d9_cl_3)' fill='#F8766D' fill-opacity='1' stroke='#F8766D' stroke-opacity='1' stroke-width='0.71' stroke-linejoin='round' stroke-linecap='round' title='<b>Species: </b>setosa'/>\n <circle cx='98.20' cy='189.90' r='1.87pt' id='svg_f97619c2-9532-4b85-acc2-030cfa5af1d9_el_24' clip-path='url(#svg_f97619c2-9532-4b85-acc2-030cfa5af1d9_cl_3)' fill='#F8766D' fill-opacity='1' stroke='#F8766D' stroke-opacity='1' stroke-width='0.71' stroke-linejoin='round' stroke-linecap='round' title='<b>Species: </b>setosa'/>\n <circle cx='83.49' cy='165.70' r='1.87pt' id='svg_f97619c2-9532-4b85-acc2-030cfa5af1d9_el_25' clip-path='url(#svg_f97619c2-9532-4b85-acc2-030cfa5af1d9_cl_3)' fill='#F8766D' fill-opacity='1' stroke='#F8766D' stroke-opacity='1' stroke-width='0.71' stroke-linejoin='round' stroke-linecap='round' title='<b>Species: </b>setosa'/>\n <circle cx='76.13' cy='177.80' r='1.87pt' id='svg_f97619c2-9532-4b85-acc2-030cfa5af1d9_el_26' clip-path='url(#svg_f97619c2-9532-4b85-acc2-030cfa5af1d9_cl_3)' fill='#F8766D' fill-opacity='1' stroke='#F8766D' stroke-opacity='1' stroke-width='0.71' stroke-linejoin='round' stroke-linecap='round' title='<b>Species: </b>setosa'/>\n <circle cx='105.55' cy='117.30' r='1.87pt' id='svg_f97619c2-9532-4b85-acc2-030cfa5af1d9_el_27' clip-path='url(#svg_f97619c2-9532-4b85-acc2-030cfa5af1d9_cl_3)' fill='#F8766D' fill-opacity='1' stroke='#F8766D' stroke-opacity='1' stroke-width='0.71' stroke-linejoin='round' stroke-linecap='round' title='<b>Species: </b>setosa'/>\n <circle cx='134.97' cy='81.00' r='1.87pt' id='svg_f97619c2-9532-4b85-acc2-030cfa5af1d9_el_28' clip-path='url(#svg_f97619c2-9532-4b85-acc2-030cfa5af1d9_cl_3)' fill='#F8766D' fill-opacity='1' stroke='#F8766D' stroke-opacity='1' stroke-width='0.71' stroke-linejoin='round' stroke-linecap='round' title='<b>Species: </b>setosa'/>\n <circle cx='76.13' cy='141.50' r='1.87pt' id='svg_f97619c2-9532-4b85-acc2-030cfa5af1d9_el_29' clip-path='url(#svg_f97619c2-9532-4b85-acc2-030cfa5af1d9_cl_3)' fill='#F8766D' fill-opacity='1' stroke='#F8766D' stroke-opacity='1' stroke-width='0.71' stroke-linejoin='round' stroke-linecap='round' title='<b>Species: </b>setosa'/>\n <circle cx='105.55' cy='141.50' r='1.87pt' id='svg_f97619c2-9532-4b85-acc2-030cfa5af1d9_el_30' clip-path='url(#svg_f97619c2-9532-4b85-acc2-030cfa5af1d9_cl_3)' fill='#F8766D' fill-opacity='1' stroke='#F8766D' stroke-opacity='1' stroke-width='0.71' stroke-linejoin='round' stroke-linecap='round' title='<b>Species: </b>setosa'/>\n <circle cx='61.43' cy='202.00' r='1.87pt' id='svg_f97619c2-9532-4b85-acc2-030cfa5af1d9_el_31' clip-path='url(#svg_f97619c2-9532-4b85-acc2-030cfa5af1d9_cl_3)' fill='#F8766D' fill-opacity='1' stroke='#F8766D' stroke-opacity='1' stroke-width='0.71' stroke-linejoin='round' stroke-linecap='round' title='<b>Species: </b>setosa'/>\n <circle cx='98.20' cy='177.80' r='1.87pt' id='svg_f97619c2-9532-4b85-acc2-030cfa5af1d9_el_32' clip-path='url(#svg_f97619c2-9532-4b85-acc2-030cfa5af1d9_cl_3)' fill='#F8766D' fill-opacity='1' stroke='#F8766D' stroke-opacity='1' stroke-width='0.71' stroke-linejoin='round' stroke-linecap='round' title='<b>Species: </b>setosa'/>\n <circle cx='134.97' cy='105.20' r='1.87pt' id='svg_f97619c2-9532-4b85-acc2-030cfa5af1d9_el_33' clip-path='url(#svg_f97619c2-9532-4b85-acc2-030cfa5af1d9_cl_3)' fill='#F8766D' fill-opacity='1' stroke='#F8766D' stroke-opacity='1' stroke-width='0.71' stroke-linejoin='round' stroke-linecap='round' title='<b>Species: </b>setosa'/>\n <circle cx='90.84' cy='141.50' r='1.87pt' id='svg_f97619c2-9532-4b85-acc2-030cfa5af1d9_el_34' clip-path='url(#svg_f97619c2-9532-4b85-acc2-030cfa5af1d9_cl_3)' fill='#F8766D' fill-opacity='1' stroke='#F8766D' stroke-opacity='1' stroke-width='0.71' stroke-linejoin='round' stroke-linecap='round' title='<b>Species: </b>setosa'/>\n <circle cx='90.84' cy='189.90' r='1.87pt' id='svg_f97619c2-9532-4b85-acc2-030cfa5af1d9_el_35' clip-path='url(#svg_f97619c2-9532-4b85-acc2-030cfa5af1d9_cl_3)' fill='#F8766D' fill-opacity='1' stroke='#F8766D' stroke-opacity='1' stroke-width='0.71' stroke-linejoin='round' stroke-linecap='round' title='<b>Species: </b>setosa'/>\n <circle cx='54.07' cy='189.90' r='1.87pt' id='svg_f97619c2-9532-4b85-acc2-030cfa5af1d9_el_36' clip-path='url(#svg_f97619c2-9532-4b85-acc2-030cfa5af1d9_cl_3)' fill='#F8766D' fill-opacity='1' stroke='#F8766D' stroke-opacity='1' stroke-width='0.71' stroke-linejoin='round' stroke-linecap='round' title='<b>Species: </b>setosa'/>\n <circle cx='164.38' cy='68.90' r='1.87pt' id='svg_f97619c2-9532-4b85-acc2-030cfa5af1d9_el_37' clip-path='url(#svg_f97619c2-9532-4b85-acc2-030cfa5af1d9_cl_3)' fill='#F8766D' fill-opacity='1' stroke='#F8766D' stroke-opacity='1' stroke-width='0.71' stroke-linejoin='round' stroke-linecap='round' title='<b>Species: </b>setosa'/>\n <circle cx='157.03' cy='20.50' r='1.87pt' id='svg_f97619c2-9532-4b85-acc2-030cfa5af1d9_el_38' clip-path='url(#svg_f97619c2-9532-4b85-acc2-030cfa5af1d9_cl_3)' fill='#F8766D' fill-opacity='1' stroke='#F8766D' stroke-opacity='1' stroke-width='0.71' stroke-linejoin='round' stroke-linecap='round' title='<b>Species: </b>setosa'/>\n <circle cx='134.97' cy='81.00' r='1.87pt' id='svg_f97619c2-9532-4b85-acc2-030cfa5af1d9_el_39' clip-path='url(#svg_f97619c2-9532-4b85-acc2-030cfa5af1d9_cl_3)' fill='#F8766D' fill-opacity='1' stroke='#F8766D' stroke-opacity='1' stroke-width='0.71' stroke-linejoin='round' stroke-linecap='round' title='<b>Species: </b>setosa'/>\n <circle cx='112.90' cy='129.40' r='1.87pt' id='svg_f97619c2-9532-4b85-acc2-030cfa5af1d9_el_40' clip-path='url(#svg_f97619c2-9532-4b85-acc2-030cfa5af1d9_cl_3)' fill='#F8766D' fill-opacity='1' stroke='#F8766D' stroke-opacity='1' stroke-width='0.71' stroke-linejoin='round' stroke-linecap='round' title='<b>Species: </b>setosa'/>\n <circle cx='157.03' cy='93.10' r='1.87pt' id='svg_f97619c2-9532-4b85-acc2-030cfa5af1d9_el_41' clip-path='url(#svg_f97619c2-9532-4b85-acc2-030cfa5af1d9_cl_3)' fill='#F8766D' fill-opacity='1' stroke='#F8766D' stroke-opacity='1' stroke-width='0.71' stroke-linejoin='round' stroke-linecap='round' title='<b>Species: </b>setosa'/>\n <circle cx='112.90' cy='93.10' r='1.87pt' id='svg_f97619c2-9532-4b85-acc2-030cfa5af1d9_el_42' clip-path='url(#svg_f97619c2-9532-4b85-acc2-030cfa5af1d9_cl_3)' fill='#F8766D' fill-opacity='1' stroke='#F8766D' stroke-opacity='1' stroke-width='0.71' stroke-linejoin='round' stroke-linecap='round' title='<b>Species: </b>setosa'/>\n <circle cx='134.97' cy='141.50' r='1.87pt' id='svg_f97619c2-9532-4b85-acc2-030cfa5af1d9_el_43' clip-path='url(#svg_f97619c2-9532-4b85-acc2-030cfa5af1d9_cl_3)' fill='#F8766D' fill-opacity='1' stroke='#F8766D' stroke-opacity='1' stroke-width='0.71' stroke-linejoin='round' stroke-linecap='round' title='<b>Species: </b>setosa'/>\n <circle cx='112.90' cy='105.20' r='1.87pt' id='svg_f97619c2-9532-4b85-acc2-030cfa5af1d9_el_44' clip-path='url(#svg_f97619c2-9532-4b85-acc2-030cfa5af1d9_cl_3)' fill='#F8766D' fill-opacity='1' stroke='#F8766D' stroke-opacity='1' stroke-width='0.71' stroke-linejoin='round' stroke-linecap='round' title='<b>Species: </b>setosa'/>\n <circle cx='76.13' cy='117.30' r='1.87pt' id='svg_f97619c2-9532-4b85-acc2-030cfa5af1d9_el_45' clip-path='url(#svg_f97619c2-9532-4b85-acc2-030cfa5af1d9_cl_3)' fill='#F8766D' fill-opacity='1' stroke='#F8766D' stroke-opacity='1' stroke-width='0.71' stroke-linejoin='round' stroke-linecap='round' title='<b>Species: </b>setosa'/>\n <circle cx='112.90' cy='153.60' r='1.87pt' id='svg_f97619c2-9532-4b85-acc2-030cfa5af1d9_el_46' clip-path='url(#svg_f97619c2-9532-4b85-acc2-030cfa5af1d9_cl_3)' fill='#F8766D' fill-opacity='1' stroke='#F8766D' stroke-opacity='1' stroke-width='0.71' stroke-linejoin='round' stroke-linecap='round' title='<b>Species: </b>setosa'/>\n <circle cx='90.84' cy='141.50' r='1.87pt' id='svg_f97619c2-9532-4b85-acc2-030cfa5af1d9_el_47' clip-path='url(#svg_f97619c2-9532-4b85-acc2-030cfa5af1d9_cl_3)' fill='#F8766D' fill-opacity='1' stroke='#F8766D' stroke-opacity='1' stroke-width='0.71' stroke-linejoin='round' stroke-linecap='round' title='<b>Species: </b>setosa'/>\n <circle cx='105.55' cy='189.90' r='1.87pt' id='svg_f97619c2-9532-4b85-acc2-030cfa5af1d9_el_48' clip-path='url(#svg_f97619c2-9532-4b85-acc2-030cfa5af1d9_cl_3)' fill='#F8766D' fill-opacity='1' stroke='#F8766D' stroke-opacity='1' stroke-width='0.71' stroke-linejoin='round' stroke-linecap='round' title='<b>Species: </b>setosa'/>\n <circle cx='105.55' cy='141.50' r='1.87pt' id='svg_f97619c2-9532-4b85-acc2-030cfa5af1d9_el_49' clip-path='url(#svg_f97619c2-9532-4b85-acc2-030cfa5af1d9_cl_3)' fill='#F8766D' fill-opacity='1' stroke='#F8766D' stroke-opacity='1' stroke-width='0.71' stroke-linejoin='round' stroke-linecap='round' title='<b>Species: </b>setosa'/>\n <circle cx='120.26' cy='129.40' r='1.87pt' id='svg_f97619c2-9532-4b85-acc2-030cfa5af1d9_el_50' clip-path='url(#svg_f97619c2-9532-4b85-acc2-030cfa5af1d9_cl_3)' fill='#F8766D' fill-opacity='1' stroke='#F8766D' stroke-opacity='1' stroke-width='0.71' stroke-linejoin='round' stroke-linecap='round' title='<b>Species: </b>setosa'/>\n <circle cx='120.26' cy='141.50' r='1.87pt' id='svg_f97619c2-9532-4b85-acc2-030cfa5af1d9_el_51' clip-path='url(#svg_f97619c2-9532-4b85-acc2-030cfa5af1d9_cl_3)' fill='#F8766D' fill-opacity='1' stroke='#F8766D' stroke-opacity='1' stroke-width='0.71' stroke-linejoin='round' stroke-linecap='round' title='<b>Species: </b>setosa'/>\n <circle cx='83.49' cy='165.70' r='1.87pt' id='svg_f97619c2-9532-4b85-acc2-030cfa5af1d9_el_52' clip-path='url(#svg_f97619c2-9532-4b85-acc2-030cfa5af1d9_cl_3)' fill='#F8766D' fill-opacity='1' stroke='#F8766D' stroke-opacity='1' stroke-width='0.71' stroke-linejoin='round' stroke-linecap='round' title='<b>Species: </b>setosa'/>\n <circle cx='90.84' cy='177.80' r='1.87pt' id='svg_f97619c2-9532-4b85-acc2-030cfa5af1d9_el_53' clip-path='url(#svg_f97619c2-9532-4b85-acc2-030cfa5af1d9_cl_3)' fill='#F8766D' fill-opacity='1' stroke='#F8766D' stroke-opacity='1' stroke-width='0.71' stroke-linejoin='round' stroke-linecap='round' title='<b>Species: </b>setosa'/>\n <circle cx='134.97' cy='141.50' r='1.87pt' id='svg_f97619c2-9532-4b85-acc2-030cfa5af1d9_el_54' clip-path='url(#svg_f97619c2-9532-4b85-acc2-030cfa5af1d9_cl_3)' fill='#F8766D' fill-opacity='1' stroke='#F8766D' stroke-opacity='1' stroke-width='0.71' stroke-linejoin='round' stroke-linecap='round' title='<b>Species: </b>setosa'/>\n <circle cx='120.26' cy='56.80' r='1.87pt' id='svg_f97619c2-9532-4b85-acc2-030cfa5af1d9_el_55' clip-path='url(#svg_f97619c2-9532-4b85-acc2-030cfa5af1d9_cl_3)' fill='#F8766D' fill-opacity='1' stroke='#F8766D' stroke-opacity='1' stroke-width='0.71' stroke-linejoin='round' stroke-linecap='round' title='<b>Species: </b>setosa'/>\n <circle cx='142.32' cy='44.70' r='1.87pt' id='svg_f97619c2-9532-4b85-acc2-030cfa5af1d9_el_56' clip-path='url(#svg_f97619c2-9532-4b85-acc2-030cfa5af1d9_cl_3)' fill='#F8766D' fill-opacity='1' stroke='#F8766D' stroke-opacity='1' stroke-width='0.71' stroke-linejoin='round' stroke-linecap='round' title='<b>Species: </b>setosa'/>\n <circle cx='98.20' cy='177.80' r='1.87pt' id='svg_f97619c2-9532-4b85-acc2-030cfa5af1d9_el_57' clip-path='url(#svg_f97619c2-9532-4b85-acc2-030cfa5af1d9_cl_3)' fill='#F8766D' fill-opacity='1' stroke='#F8766D' stroke-opacity='1' stroke-width='0.71' stroke-linejoin='round' stroke-linecap='round' title='<b>Species: </b>setosa'/>\n <circle cx='105.55' cy='165.70' r='1.87pt' id='svg_f97619c2-9532-4b85-acc2-030cfa5af1d9_el_58' clip-path='url(#svg_f97619c2-9532-4b85-acc2-030cfa5af1d9_cl_3)' fill='#F8766D' fill-opacity='1' stroke='#F8766D' stroke-opacity='1' stroke-width='0.71' stroke-linejoin='round' stroke-linecap='round' title='<b>Species: </b>setosa'/>\n <circle cx='142.32' cy='129.40' r='1.87pt' id='svg_f97619c2-9532-4b85-acc2-030cfa5af1d9_el_59' clip-path='url(#svg_f97619c2-9532-4b85-acc2-030cfa5af1d9_cl_3)' fill='#F8766D' fill-opacity='1' stroke='#F8766D' stroke-opacity='1' stroke-width='0.71' stroke-linejoin='round' stroke-linecap='round' title='<b>Species: </b>setosa'/>\n <circle cx='98.20' cy='117.30' r='1.87pt' id='svg_f97619c2-9532-4b85-acc2-030cfa5af1d9_el_60' clip-path='url(#svg_f97619c2-9532-4b85-acc2-030cfa5af1d9_cl_3)' fill='#F8766D' fill-opacity='1' stroke='#F8766D' stroke-opacity='1' stroke-width='0.71' stroke-linejoin='round' stroke-linecap='round' title='<b>Species: </b>setosa'/>\n <circle cx='61.43' cy='189.90' r='1.87pt' id='svg_f97619c2-9532-4b85-acc2-030cfa5af1d9_el_61' clip-path='url(#svg_f97619c2-9532-4b85-acc2-030cfa5af1d9_cl_3)' fill='#F8766D' fill-opacity='1' stroke='#F8766D' stroke-opacity='1' stroke-width='0.71' stroke-linejoin='round' stroke-linecap='round' title='<b>Species: </b>setosa'/>\n <circle cx='112.90' cy='141.50' r='1.87pt' id='svg_f97619c2-9532-4b85-acc2-030cfa5af1d9_el_62' clip-path='url(#svg_f97619c2-9532-4b85-acc2-030cfa5af1d9_cl_3)' fill='#F8766D' fill-opacity='1' stroke='#F8766D' stroke-opacity='1' stroke-width='0.71' stroke-linejoin='round' stroke-linecap='round' title='<b>Species: </b>setosa'/>\n <circle cx='105.55' cy='129.40' r='1.87pt' id='svg_f97619c2-9532-4b85-acc2-030cfa5af1d9_el_63' clip-path='url(#svg_f97619c2-9532-4b85-acc2-030cfa5af1d9_cl_3)' fill='#F8766D' fill-opacity='1' stroke='#F8766D' stroke-opacity='1' stroke-width='0.71' stroke-linejoin='round' stroke-linecap='round' title='<b>Species: </b>setosa'/>\n <circle cx='68.78' cy='274.60' r='1.87pt' id='svg_f97619c2-9532-4b85-acc2-030cfa5af1d9_el_64' clip-path='url(#svg_f97619c2-9532-4b85-acc2-030cfa5af1d9_cl_3)' fill='#F8766D' fill-opacity='1' stroke='#F8766D' stroke-opacity='1' stroke-width='0.71' stroke-linejoin='round' stroke-linecap='round' title='<b>Species: </b>setosa'/>\n <circle cx='61.43' cy='165.70' r='1.87pt' id='svg_f97619c2-9532-4b85-acc2-030cfa5af1d9_el_65' clip-path='url(#svg_f97619c2-9532-4b85-acc2-030cfa5af1d9_cl_3)' fill='#F8766D' fill-opacity='1' stroke='#F8766D' stroke-opacity='1' stroke-width='0.71' stroke-linejoin='round' stroke-linecap='round' title='<b>Species: </b>setosa'/>\n <circle cx='105.55' cy='129.40' r='1.87pt' id='svg_f97619c2-9532-4b85-acc2-030cfa5af1d9_el_66' clip-path='url(#svg_f97619c2-9532-4b85-acc2-030cfa5af1d9_cl_3)' fill='#F8766D' fill-opacity='1' stroke='#F8766D' stroke-opacity='1' stroke-width='0.71' stroke-linejoin='round' stroke-linecap='round' title='<b>Species: </b>setosa'/>\n <circle cx='112.90' cy='93.10' r='1.87pt' id='svg_f97619c2-9532-4b85-acc2-030cfa5af1d9_el_67' clip-path='url(#svg_f97619c2-9532-4b85-acc2-030cfa5af1d9_cl_3)' fill='#F8766D' fill-opacity='1' stroke='#F8766D' stroke-opacity='1' stroke-width='0.71' stroke-linejoin='round' stroke-linecap='round' title='<b>Species: </b>setosa'/>\n <circle cx='90.84' cy='189.90' r='1.87pt' id='svg_f97619c2-9532-4b85-acc2-030cfa5af1d9_el_68' clip-path='url(#svg_f97619c2-9532-4b85-acc2-030cfa5af1d9_cl_3)' fill='#F8766D' fill-opacity='1' stroke='#F8766D' stroke-opacity='1' stroke-width='0.71' stroke-linejoin='round' stroke-linecap='round' title='<b>Species: </b>setosa'/>\n <circle cx='112.90' cy='93.10' r='1.87pt' id='svg_f97619c2-9532-4b85-acc2-030cfa5af1d9_el_69' clip-path='url(#svg_f97619c2-9532-4b85-acc2-030cfa5af1d9_cl_3)' fill='#F8766D' fill-opacity='1' stroke='#F8766D' stroke-opacity='1' stroke-width='0.71' stroke-linejoin='round' stroke-linecap='round' title='<b>Species: </b>setosa'/>\n <circle cx='76.13' cy='165.70' r='1.87pt' id='svg_f97619c2-9532-4b85-acc2-030cfa5af1d9_el_70' clip-path='url(#svg_f97619c2-9532-4b85-acc2-030cfa5af1d9_cl_3)' fill='#F8766D' fill-opacity='1' stroke='#F8766D' stroke-opacity='1' stroke-width='0.71' stroke-linejoin='round' stroke-linecap='round' title='<b>Species: </b>setosa'/>\n <circle cx='127.61' cy='105.20' r='1.87pt' id='svg_f97619c2-9532-4b85-acc2-030cfa5af1d9_el_71' clip-path='url(#svg_f97619c2-9532-4b85-acc2-030cfa5af1d9_cl_3)' fill='#F8766D' fill-opacity='1' stroke='#F8766D' stroke-opacity='1' stroke-width='0.71' stroke-linejoin='round' stroke-linecap='round' title='<b>Species: </b>setosa'/>\n <circle cx='105.55' cy='153.60' r='1.87pt' id='svg_f97619c2-9532-4b85-acc2-030cfa5af1d9_el_72' clip-path='url(#svg_f97619c2-9532-4b85-acc2-030cfa5af1d9_cl_3)' fill='#F8766D' fill-opacity='1' stroke='#F8766D' stroke-opacity='1' stroke-width='0.71' stroke-linejoin='round' stroke-linecap='round' title='<b>Species: </b>setosa'/>\n <circle cx='252.63' cy='165.70' r='1.87pt' id='svg_f97619c2-9532-4b85-acc2-030cfa5af1d9_el_73' clip-path='url(#svg_f97619c2-9532-4b85-acc2-030cfa5af1d9_cl_3)' fill='#00BA38' fill-opacity='1' stroke='#00BA38' stroke-opacity='1' stroke-width='0.71' stroke-linejoin='round' stroke-linecap='round' title='<b>Species: </b>versicolor'/>\n <circle cx='208.51' cy='165.70' r='1.87pt' id='svg_f97619c2-9532-4b85-acc2-030cfa5af1d9_el_74' clip-path='url(#svg_f97619c2-9532-4b85-acc2-030cfa5af1d9_cl_3)' fill='#00BA38' fill-opacity='1' stroke='#00BA38' stroke-opacity='1' stroke-width='0.71' stroke-linejoin='round' stroke-linecap='round' title='<b>Species: </b>versicolor'/>\n <circle cx='245.28' cy='177.80' r='1.87pt' id='svg_f97619c2-9532-4b85-acc2-030cfa5af1d9_el_75' clip-path='url(#svg_f97619c2-9532-4b85-acc2-030cfa5af1d9_cl_3)' fill='#00BA38' fill-opacity='1' stroke='#00BA38' stroke-opacity='1' stroke-width='0.71' stroke-linejoin='round' stroke-linecap='round' title='<b>Species: </b>versicolor'/>\n <circle cx='142.32' cy='274.60' r='1.87pt' id='svg_f97619c2-9532-4b85-acc2-030cfa5af1d9_el_76' clip-path='url(#svg_f97619c2-9532-4b85-acc2-030cfa5af1d9_cl_3)' fill='#00BA38' fill-opacity='1' stroke='#00BA38' stroke-opacity='1' stroke-width='0.71' stroke-linejoin='round' stroke-linecap='round' title='<b>Species: </b>versicolor'/>\n <circle cx='215.86' cy='214.10' r='1.87pt' id='svg_f97619c2-9532-4b85-acc2-030cfa5af1d9_el_77' clip-path='url(#svg_f97619c2-9532-4b85-acc2-030cfa5af1d9_cl_3)' fill='#00BA38' fill-opacity='1' stroke='#00BA38' stroke-opacity='1' stroke-width='0.71' stroke-linejoin='round' stroke-linecap='round' title='<b>Species: </b>versicolor'/>\n <circle cx='157.03' cy='214.10' r='1.87pt' id='svg_f97619c2-9532-4b85-acc2-030cfa5af1d9_el_78' clip-path='url(#svg_f97619c2-9532-4b85-acc2-030cfa5af1d9_cl_3)' fill='#00BA38' fill-opacity='1' stroke='#00BA38' stroke-opacity='1' stroke-width='0.71' stroke-linejoin='round' stroke-linecap='round' title='<b>Species: </b>versicolor'/>\n <circle cx='201.15' cy='153.60' r='1.87pt' id='svg_f97619c2-9532-4b85-acc2-030cfa5af1d9_el_79' clip-path='url(#svg_f97619c2-9532-4b85-acc2-030cfa5af1d9_cl_3)' fill='#00BA38' fill-opacity='1' stroke='#00BA38' stroke-opacity='1' stroke-width='0.71' stroke-linejoin='round' stroke-linecap='round' title='<b>Species: </b>versicolor'/>\n <circle cx='98.20' cy='262.50' r='1.87pt' id='svg_f97619c2-9532-4b85-acc2-030cfa5af1d9_el_80' clip-path='url(#svg_f97619c2-9532-4b85-acc2-030cfa5af1d9_cl_3)' fill='#00BA38' fill-opacity='1' stroke='#00BA38' stroke-opacity='1' stroke-width='0.71' stroke-linejoin='round' stroke-linecap='round' title='<b>Species: </b>versicolor'/>\n <circle cx='223.21' cy='202.00' r='1.87pt' id='svg_f97619c2-9532-4b85-acc2-030cfa5af1d9_el_81' clip-path='url(#svg_f97619c2-9532-4b85-acc2-030cfa5af1d9_cl_3)' fill='#00BA38' fill-opacity='1' stroke='#00BA38' stroke-opacity='1' stroke-width='0.71' stroke-linejoin='round' stroke-linecap='round' title='<b>Species: </b>versicolor'/>\n <circle cx='120.26' cy='226.20' r='1.87pt' id='svg_f97619c2-9532-4b85-acc2-030cfa5af1d9_el_82' clip-path='url(#svg_f97619c2-9532-4b85-acc2-030cfa5af1d9_cl_3)' fill='#00BA38' fill-opacity='1' stroke='#00BA38' stroke-opacity='1' stroke-width='0.71' stroke-linejoin='round' stroke-linecap='round' title='<b>Species: </b>versicolor'/>\n <circle cx='105.55' cy='310.90' r='1.87pt' id='svg_f97619c2-9532-4b85-acc2-030cfa5af1d9_el_83' clip-path='url(#svg_f97619c2-9532-4b85-acc2-030cfa5af1d9_cl_3)' fill='#00BA38' fill-opacity='1' stroke='#00BA38' stroke-opacity='1' stroke-width='0.71' stroke-linejoin='round' stroke-linecap='round' title='<b>Species: </b>versicolor'/>\n <circle cx='171.74' cy='189.90' r='1.87pt' id='svg_f97619c2-9532-4b85-acc2-030cfa5af1d9_el_84' clip-path='url(#svg_f97619c2-9532-4b85-acc2-030cfa5af1d9_cl_3)' fill='#00BA38' fill-opacity='1' stroke='#00BA38' stroke-opacity='1' stroke-width='0.71' stroke-linejoin='round' stroke-linecap='round' title='<b>Species: </b>versicolor'/>\n <circle cx='179.09' cy='286.70' r='1.87pt' id='svg_f97619c2-9532-4b85-acc2-030cfa5af1d9_el_85' clip-path='url(#svg_f97619c2-9532-4b85-acc2-030cfa5af1d9_cl_3)' fill='#00BA38' fill-opacity='1' stroke='#00BA38' stroke-opacity='1' stroke-width='0.71' stroke-linejoin='round' stroke-linecap='round' title='<b>Species: </b>versicolor'/>\n <circle cx='186.44' cy='202.00' r='1.87pt' id='svg_f97619c2-9532-4b85-acc2-030cfa5af1d9_el_86' clip-path='url(#svg_f97619c2-9532-4b85-acc2-030cfa5af1d9_cl_3)' fill='#00BA38' fill-opacity='1' stroke='#00BA38' stroke-opacity='1' stroke-width='0.71' stroke-linejoin='round' stroke-linecap='round' title='<b>Species: </b>versicolor'/>\n <circle cx='149.67' cy='202.00' r='1.87pt' id='svg_f97619c2-9532-4b85-acc2-030cfa5af1d9_el_87' clip-path='url(#svg_f97619c2-9532-4b85-acc2-030cfa5af1d9_cl_3)' fill='#00BA38' fill-opacity='1' stroke='#00BA38' stroke-opacity='1' stroke-width='0.71' stroke-linejoin='round' stroke-linecap='round' title='<b>Species: </b>versicolor'/>\n <circle cx='230.57' cy='177.80' r='1.87pt' id='svg_f97619c2-9532-4b85-acc2-030cfa5af1d9_el_88' clip-path='url(#svg_f97619c2-9532-4b85-acc2-030cfa5af1d9_cl_3)' fill='#00BA38' fill-opacity='1' stroke='#00BA38' stroke-opacity='1' stroke-width='0.71' stroke-linejoin='round' stroke-linecap='round' title='<b>Species: </b>versicolor'/>\n <circle cx='149.67' cy='189.90' r='1.87pt' id='svg_f97619c2-9532-4b85-acc2-030cfa5af1d9_el_89' clip-path='url(#svg_f97619c2-9532-4b85-acc2-030cfa5af1d9_cl_3)' fill='#00BA38' fill-opacity='1' stroke='#00BA38' stroke-opacity='1' stroke-width='0.71' stroke-linejoin='round' stroke-linecap='round' title='<b>Species: </b>versicolor'/>\n <circle cx='164.38' cy='226.20' r='1.87pt' id='svg_f97619c2-9532-4b85-acc2-030cfa5af1d9_el_90' clip-path='url(#svg_f97619c2-9532-4b85-acc2-030cfa5af1d9_cl_3)' fill='#00BA38' fill-opacity='1' stroke='#00BA38' stroke-opacity='1' stroke-width='0.71' stroke-linejoin='round' stroke-linecap='round' title='<b>Species: </b>versicolor'/>\n <circle cx='193.80' cy='286.70' r='1.87pt' id='svg_f97619c2-9532-4b85-acc2-030cfa5af1d9_el_91' clip-path='url(#svg_f97619c2-9532-4b85-acc2-030cfa5af1d9_cl_3)' fill='#00BA38' fill-opacity='1' stroke='#00BA38' stroke-opacity='1' stroke-width='0.71' stroke-linejoin='round' stroke-linecap='round' title='<b>Species: </b>versicolor'/>\n <circle cx='149.67' cy='250.40' r='1.87pt' id='svg_f97619c2-9532-4b85-acc2-030cfa5af1d9_el_92' clip-path='url(#svg_f97619c2-9532-4b85-acc2-030cfa5af1d9_cl_3)' fill='#00BA38' fill-opacity='1' stroke='#00BA38' stroke-opacity='1' stroke-width='0.71' stroke-linejoin='round' stroke-linecap='round' title='<b>Species: </b>versicolor'/>\n <circle cx='171.74' cy='165.70' r='1.87pt' id='svg_f97619c2-9532-4b85-acc2-030cfa5af1d9_el_93' clip-path='url(#svg_f97619c2-9532-4b85-acc2-030cfa5af1d9_cl_3)' fill='#00BA38' fill-opacity='1' stroke='#00BA38' stroke-opacity='1' stroke-width='0.71' stroke-linejoin='round' stroke-linecap='round' title='<b>Species: </b>versicolor'/>\n <circle cx='186.44' cy='214.10' r='1.87pt' id='svg_f97619c2-9532-4b85-acc2-030cfa5af1d9_el_94' clip-path='url(#svg_f97619c2-9532-4b85-acc2-030cfa5af1d9_cl_3)' fill='#00BA38' fill-opacity='1' stroke='#00BA38' stroke-opacity='1' stroke-width='0.71' stroke-linejoin='round' stroke-linecap='round' title='<b>Species: </b>versicolor'/>\n <circle cx='201.15' cy='250.40' r='1.87pt' id='svg_f97619c2-9532-4b85-acc2-030cfa5af1d9_el_95' clip-path='url(#svg_f97619c2-9532-4b85-acc2-030cfa5af1d9_cl_3)' fill='#00BA38' fill-opacity='1' stroke='#00BA38' stroke-opacity='1' stroke-width='0.71' stroke-linejoin='round' stroke-linecap='round' title='<b>Species: </b>versicolor'/>\n <circle cx='186.44' cy='214.10' r='1.87pt' id='svg_f97619c2-9532-4b85-acc2-030cfa5af1d9_el_96' clip-path='url(#svg_f97619c2-9532-4b85-acc2-030cfa5af1d9_cl_3)' fill='#00BA38' fill-opacity='1' stroke='#00BA38' stroke-opacity='1' stroke-width='0.71' stroke-linejoin='round' stroke-linecap='round' title='<b>Species: </b>versicolor'/>\n <circle cx='208.51' cy='202.00' r='1.87pt' id='svg_f97619c2-9532-4b85-acc2-030cfa5af1d9_el_97' clip-path='url(#svg_f97619c2-9532-4b85-acc2-030cfa5af1d9_cl_3)' fill='#00BA38' fill-opacity='1' stroke='#00BA38' stroke-opacity='1' stroke-width='0.71' stroke-linejoin='round' stroke-linecap='round' title='<b>Species: </b>versicolor'/>\n <circle cx='223.21' cy='189.90' r='1.87pt' id='svg_f97619c2-9532-4b85-acc2-030cfa5af1d9_el_98' clip-path='url(#svg_f97619c2-9532-4b85-acc2-030cfa5af1d9_cl_3)' fill='#00BA38' fill-opacity='1' stroke='#00BA38' stroke-opacity='1' stroke-width='0.71' stroke-linejoin='round' stroke-linecap='round' title='<b>Species: </b>versicolor'/>\n <circle cx='237.92' cy='214.10' r='1.87pt' id='svg_f97619c2-9532-4b85-acc2-030cfa5af1d9_el_99' clip-path='url(#svg_f97619c2-9532-4b85-acc2-030cfa5af1d9_cl_3)' fill='#00BA38' fill-opacity='1' stroke='#00BA38' stroke-opacity='1' stroke-width='0.71' stroke-linejoin='round' stroke-linecap='round' title='<b>Species: </b>versicolor'/>\n <circle cx='230.57' cy='189.90' r='1.87pt' id='svg_f97619c2-9532-4b85-acc2-030cfa5af1d9_el_100' clip-path='url(#svg_f97619c2-9532-4b85-acc2-030cfa5af1d9_cl_3)' fill='#00BA38' fill-opacity='1' stroke='#00BA38' stroke-opacity='1' stroke-width='0.71' stroke-linejoin='round' stroke-linecap='round' title='<b>Species: </b>versicolor'/>\n <circle cx='179.09' cy='202.00' r='1.87pt' id='svg_f97619c2-9532-4b85-acc2-030cfa5af1d9_el_101' clip-path='url(#svg_f97619c2-9532-4b85-acc2-030cfa5af1d9_cl_3)' fill='#00BA38' fill-opacity='1' stroke='#00BA38' stroke-opacity='1' stroke-width='0.71' stroke-linejoin='round' stroke-linecap='round' title='<b>Species: </b>versicolor'/>\n <circle cx='157.03' cy='238.30' r='1.87pt' id='svg_f97619c2-9532-4b85-acc2-030cfa5af1d9_el_102' clip-path='url(#svg_f97619c2-9532-4b85-acc2-030cfa5af1d9_cl_3)' fill='#00BA38' fill-opacity='1' stroke='#00BA38' stroke-opacity='1' stroke-width='0.71' stroke-linejoin='round' stroke-linecap='round' title='<b>Species: </b>versicolor'/>\n <circle cx='142.32' cy='262.50' r='1.87pt' id='svg_f97619c2-9532-4b85-acc2-030cfa5af1d9_el_103' clip-path='url(#svg_f97619c2-9532-4b85-acc2-030cfa5af1d9_cl_3)' fill='#00BA38' fill-opacity='1' stroke='#00BA38' stroke-opacity='1' stroke-width='0.71' stroke-linejoin='round' stroke-linecap='round' title='<b>Species: </b>versicolor'/>\n <circle cx='142.32' cy='262.50' r='1.87pt' id='svg_f97619c2-9532-4b85-acc2-030cfa5af1d9_el_104' clip-path='url(#svg_f97619c2-9532-4b85-acc2-030cfa5af1d9_cl_3)' fill='#00BA38' fill-opacity='1' stroke='#00BA38' stroke-opacity='1' stroke-width='0.71' stroke-linejoin='round' stroke-linecap='round' title='<b>Species: </b>versicolor'/>\n <circle cx='164.38' cy='226.20' r='1.87pt' id='svg_f97619c2-9532-4b85-acc2-030cfa5af1d9_el_105' clip-path='url(#svg_f97619c2-9532-4b85-acc2-030cfa5af1d9_cl_3)' fill='#00BA38' fill-opacity='1' stroke='#00BA38' stroke-opacity='1' stroke-width='0.71' stroke-linejoin='round' stroke-linecap='round' title='<b>Species: </b>versicolor'/>\n <circle cx='179.09' cy='226.20' r='1.87pt' id='svg_f97619c2-9532-4b85-acc2-030cfa5af1d9_el_106' clip-path='url(#svg_f97619c2-9532-4b85-acc2-030cfa5af1d9_cl_3)' fill='#00BA38' fill-opacity='1' stroke='#00BA38' stroke-opacity='1' stroke-width='0.71' stroke-linejoin='round' stroke-linecap='round' title='<b>Species: </b>versicolor'/>\n <circle cx='134.97' cy='189.90' r='1.87pt' id='svg_f97619c2-9532-4b85-acc2-030cfa5af1d9_el_107' clip-path='url(#svg_f97619c2-9532-4b85-acc2-030cfa5af1d9_cl_3)' fill='#00BA38' fill-opacity='1' stroke='#00BA38' stroke-opacity='1' stroke-width='0.71' stroke-linejoin='round' stroke-linecap='round' title='<b>Species: </b>versicolor'/>\n <circle cx='179.09' cy='141.50' r='1.87pt' id='svg_f97619c2-9532-4b85-acc2-030cfa5af1d9_el_108' clip-path='url(#svg_f97619c2-9532-4b85-acc2-030cfa5af1d9_cl_3)' fill='#00BA38' fill-opacity='1' stroke='#00BA38' stroke-opacity='1' stroke-width='0.71' stroke-linejoin='round' stroke-linecap='round' title='<b>Species: </b>versicolor'/>\n <circle cx='230.57' cy='177.80' r='1.87pt' id='svg_f97619c2-9532-4b85-acc2-030cfa5af1d9_el_109' clip-path='url(#svg_f97619c2-9532-4b85-acc2-030cfa5af1d9_cl_3)' fill='#00BA38' fill-opacity='1' stroke='#00BA38' stroke-opacity='1' stroke-width='0.71' stroke-linejoin='round' stroke-linecap='round' title='<b>Species: </b>versicolor'/>\n <circle cx='201.15' cy='274.60' r='1.87pt' id='svg_f97619c2-9532-4b85-acc2-030cfa5af1d9_el_110' clip-path='url(#svg_f97619c2-9532-4b85-acc2-030cfa5af1d9_cl_3)' fill='#00BA38' fill-opacity='1' stroke='#00BA38' stroke-opacity='1' stroke-width='0.71' stroke-linejoin='round' stroke-linecap='round' title='<b>Species: </b>versicolor'/>\n <circle cx='149.67' cy='189.90' r='1.87pt' id='svg_f97619c2-9532-4b85-acc2-030cfa5af1d9_el_111' clip-path='url(#svg_f97619c2-9532-4b85-acc2-030cfa5af1d9_cl_3)' fill='#00BA38' fill-opacity='1' stroke='#00BA38' stroke-opacity='1' stroke-width='0.71' stroke-linejoin='round' stroke-linecap='round' title='<b>Species: </b>versicolor'/>\n <circle cx='142.32' cy='250.40' r='1.87pt' id='svg_f97619c2-9532-4b85-acc2-030cfa5af1d9_el_112' clip-path='url(#svg_f97619c2-9532-4b85-acc2-030cfa5af1d9_cl_3)' fill='#00BA38' fill-opacity='1' stroke='#00BA38' stroke-opacity='1' stroke-width='0.71' stroke-linejoin='round' stroke-linecap='round' title='<b>Species: </b>versicolor'/>\n <circle cx='142.32' cy='238.30' r='1.87pt' id='svg_f97619c2-9532-4b85-acc2-030cfa5af1d9_el_113' clip-path='url(#svg_f97619c2-9532-4b85-acc2-030cfa5af1d9_cl_3)' fill='#00BA38' fill-opacity='1' stroke='#00BA38' stroke-opacity='1' stroke-width='0.71' stroke-linejoin='round' stroke-linecap='round' title='<b>Species: </b>versicolor'/>\n <circle cx='186.44' cy='189.90' r='1.87pt' id='svg_f97619c2-9532-4b85-acc2-030cfa5af1d9_el_114' clip-path='url(#svg_f97619c2-9532-4b85-acc2-030cfa5af1d9_cl_3)' fill='#00BA38' fill-opacity='1' stroke='#00BA38' stroke-opacity='1' stroke-width='0.71' stroke-linejoin='round' stroke-linecap='round' title='<b>Species: </b>versicolor'/>\n <circle cx='164.38' cy='238.30' r='1.87pt' id='svg_f97619c2-9532-4b85-acc2-030cfa5af1d9_el_115' clip-path='url(#svg_f97619c2-9532-4b85-acc2-030cfa5af1d9_cl_3)' fill='#00BA38' fill-opacity='1' stroke='#00BA38' stroke-opacity='1' stroke-width='0.71' stroke-linejoin='round' stroke-linecap='round' title='<b>Species: </b>versicolor'/>\n <circle cx='105.55' cy='274.60' r='1.87pt' id='svg_f97619c2-9532-4b85-acc2-030cfa5af1d9_el_116' clip-path='url(#svg_f97619c2-9532-4b85-acc2-030cfa5af1d9_cl_3)' fill='#00BA38' fill-opacity='1' stroke='#00BA38' stroke-opacity='1' stroke-width='0.71' stroke-linejoin='round' stroke-linecap='round' title='<b>Species: </b>versicolor'/>\n <circle cx='149.67' cy='226.20' r='1.87pt' id='svg_f97619c2-9532-4b85-acc2-030cfa5af1d9_el_117' clip-path='url(#svg_f97619c2-9532-4b85-acc2-030cfa5af1d9_cl_3)' fill='#00BA38' fill-opacity='1' stroke='#00BA38' stroke-opacity='1' stroke-width='0.71' stroke-linejoin='round' stroke-linecap='round' title='<b>Species: </b>versicolor'/>\n <circle cx='157.03' cy='189.90' r='1.87pt' id='svg_f97619c2-9532-4b85-acc2-030cfa5af1d9_el_118' clip-path='url(#svg_f97619c2-9532-4b85-acc2-030cfa5af1d9_cl_3)' fill='#00BA38' fill-opacity='1' stroke='#00BA38' stroke-opacity='1' stroke-width='0.71' stroke-linejoin='round' stroke-linecap='round' title='<b>Species: </b>versicolor'/>\n <circle cx='157.03' cy='202.00' r='1.87pt' id='svg_f97619c2-9532-4b85-acc2-030cfa5af1d9_el_119' clip-path='url(#svg_f97619c2-9532-4b85-acc2-030cfa5af1d9_cl_3)' fill='#00BA38' fill-opacity='1' stroke='#00BA38' stroke-opacity='1' stroke-width='0.71' stroke-linejoin='round' stroke-linecap='round' title='<b>Species: </b>versicolor'/>\n <circle cx='193.80' cy='202.00' r='1.87pt' id='svg_f97619c2-9532-4b85-acc2-030cfa5af1d9_el_120' clip-path='url(#svg_f97619c2-9532-4b85-acc2-030cfa5af1d9_cl_3)' fill='#00BA38' fill-opacity='1' stroke='#00BA38' stroke-opacity='1' stroke-width='0.71' stroke-linejoin='round' stroke-linecap='round' title='<b>Species: </b>versicolor'/>\n <circle cx='112.90' cy='250.40' r='1.87pt' id='svg_f97619c2-9532-4b85-acc2-030cfa5af1d9_el_121' clip-path='url(#svg_f97619c2-9532-4b85-acc2-030cfa5af1d9_cl_3)' fill='#00BA38' fill-opacity='1' stroke='#00BA38' stroke-opacity='1' stroke-width='0.71' stroke-linejoin='round' stroke-linecap='round' title='<b>Species: </b>versicolor'/>\n <circle cx='157.03' cy='214.10' r='1.87pt' id='svg_f97619c2-9532-4b85-acc2-030cfa5af1d9_el_122' clip-path='url(#svg_f97619c2-9532-4b85-acc2-030cfa5af1d9_cl_3)' fill='#00BA38' fill-opacity='1' stroke='#00BA38' stroke-opacity='1' stroke-width='0.71' stroke-linejoin='round' stroke-linecap='round' title='<b>Species: </b>versicolor'/>\n <circle cx='201.15' cy='153.60' r='1.87pt' id='svg_f97619c2-9532-4b85-acc2-030cfa5af1d9_el_123' clip-path='url(#svg_f97619c2-9532-4b85-acc2-030cfa5af1d9_cl_3)' fill='#619CFF' fill-opacity='1' stroke='#619CFF' stroke-opacity='1' stroke-width='0.71' stroke-linejoin='round' stroke-linecap='round' title='<b>Species: </b>virginica'/>\n <circle cx='164.38' cy='226.20' r='1.87pt' id='svg_f97619c2-9532-4b85-acc2-030cfa5af1d9_el_124' clip-path='url(#svg_f97619c2-9532-4b85-acc2-030cfa5af1d9_cl_3)' fill='#619CFF' fill-opacity='1' stroke='#619CFF' stroke-opacity='1' stroke-width='0.71' stroke-linejoin='round' stroke-linecap='round' title='<b>Species: </b>virginica'/>\n <circle cx='259.99' cy='189.90' r='1.87pt' id='svg_f97619c2-9532-4b85-acc2-030cfa5af1d9_el_125' clip-path='url(#svg_f97619c2-9532-4b85-acc2-030cfa5af1d9_cl_3)' fill='#619CFF' fill-opacity='1' stroke='#619CFF' stroke-opacity='1' stroke-width='0.71' stroke-linejoin='round' stroke-linecap='round' title='<b>Species: </b>virginica'/>\n <circle cx='201.15' cy='202.00' r='1.87pt' id='svg_f97619c2-9532-4b85-acc2-030cfa5af1d9_el_126' clip-path='url(#svg_f97619c2-9532-4b85-acc2-030cfa5af1d9_cl_3)' fill='#619CFF' fill-opacity='1' stroke='#619CFF' stroke-opacity='1' stroke-width='0.71' stroke-linejoin='round' stroke-linecap='round' title='<b>Species: </b>virginica'/>\n <circle cx='215.86' cy='189.90' r='1.87pt' id='svg_f97619c2-9532-4b85-acc2-030cfa5af1d9_el_127' clip-path='url(#svg_f97619c2-9532-4b85-acc2-030cfa5af1d9_cl_3)' fill='#619CFF' fill-opacity='1' stroke='#619CFF' stroke-opacity='1' stroke-width='0.71' stroke-linejoin='round' stroke-linecap='round' title='<b>Species: </b>virginica'/>\n <circle cx='296.76' cy='189.90' r='1.87pt' id='svg_f97619c2-9532-4b85-acc2-030cfa5af1d9_el_128' clip-path='url(#svg_f97619c2-9532-4b85-acc2-030cfa5af1d9_cl_3)' fill='#619CFF' fill-opacity='1' stroke='#619CFF' stroke-opacity='1' stroke-width='0.71' stroke-linejoin='round' stroke-linecap='round' title='<b>Species: </b>virginica'/>\n <circle cx='98.20' cy='250.40' r='1.87pt' id='svg_f97619c2-9532-4b85-acc2-030cfa5af1d9_el_129' clip-path='url(#svg_f97619c2-9532-4b85-acc2-030cfa5af1d9_cl_3)' fill='#619CFF' fill-opacity='1' stroke='#619CFF' stroke-opacity='1' stroke-width='0.71' stroke-linejoin='round' stroke-linecap='round' title='<b>Species: </b>virginica'/>\n <circle cx='274.69' cy='202.00' r='1.87pt' id='svg_f97619c2-9532-4b85-acc2-030cfa5af1d9_el_130' clip-path='url(#svg_f97619c2-9532-4b85-acc2-030cfa5af1d9_cl_3)' fill='#619CFF' fill-opacity='1' stroke='#619CFF' stroke-opacity='1' stroke-width='0.71' stroke-linejoin='round' stroke-linecap='round' title='<b>Species: </b>virginica'/>\n <circle cx='230.57' cy='250.40' r='1.87pt' id='svg_f97619c2-9532-4b85-acc2-030cfa5af1d9_el_131' clip-path='url(#svg_f97619c2-9532-4b85-acc2-030cfa5af1d9_cl_3)' fill='#619CFF' fill-opacity='1' stroke='#619CFF' stroke-opacity='1' stroke-width='0.71' stroke-linejoin='round' stroke-linecap='round' title='<b>Species: </b>virginica'/>\n <circle cx='267.34' cy='117.30' r='1.87pt' id='svg_f97619c2-9532-4b85-acc2-030cfa5af1d9_el_132' clip-path='url(#svg_f97619c2-9532-4b85-acc2-030cfa5af1d9_cl_3)' fill='#619CFF' fill-opacity='1' stroke='#619CFF' stroke-opacity='1' stroke-width='0.71' stroke-linejoin='round' stroke-linecap='round' title='<b>Species: </b>virginica'/>\n <circle cx='215.86' cy='165.70' r='1.87pt' id='svg_f97619c2-9532-4b85-acc2-030cfa5af1d9_el_133' clip-path='url(#svg_f97619c2-9532-4b85-acc2-030cfa5af1d9_cl_3)' fill='#619CFF' fill-opacity='1' stroke='#619CFF' stroke-opacity='1' stroke-width='0.71' stroke-linejoin='round' stroke-linecap='round' title='<b>Species: </b>virginica'/>\n <circle cx='208.51' cy='226.20' r='1.87pt' id='svg_f97619c2-9532-4b85-acc2-030cfa5af1d9_el_134' clip-path='url(#svg_f97619c2-9532-4b85-acc2-030cfa5af1d9_cl_3)' fill='#619CFF' fill-opacity='1' stroke='#619CFF' stroke-opacity='1' stroke-width='0.71' stroke-linejoin='round' stroke-linecap='round' title='<b>Species: </b>virginica'/>\n <circle cx='237.92' cy='189.90' r='1.87pt' id='svg_f97619c2-9532-4b85-acc2-030cfa5af1d9_el_135' clip-path='url(#svg_f97619c2-9532-4b85-acc2-030cfa5af1d9_cl_3)' fill='#619CFF' fill-opacity='1' stroke='#619CFF' stroke-opacity='1' stroke-width='0.71' stroke-linejoin='round' stroke-linecap='round' title='<b>Species: </b>virginica'/>\n <circle cx='157.03' cy='250.40' r='1.87pt' id='svg_f97619c2-9532-4b85-acc2-030cfa5af1d9_el_136' clip-path='url(#svg_f97619c2-9532-4b85-acc2-030cfa5af1d9_cl_3)' fill='#619CFF' fill-opacity='1' stroke='#619CFF' stroke-opacity='1' stroke-width='0.71' stroke-linejoin='round' stroke-linecap='round' title='<b>Species: </b>virginica'/>\n <circle cx='164.38' cy='214.10' r='1.87pt' id='svg_f97619c2-9532-4b85-acc2-030cfa5af1d9_el_137' clip-path='url(#svg_f97619c2-9532-4b85-acc2-030cfa5af1d9_cl_3)' fill='#619CFF' fill-opacity='1' stroke='#619CFF' stroke-opacity='1' stroke-width='0.71' stroke-linejoin='round' stroke-linecap='round' title='<b>Species: </b>virginica'/>\n <circle cx='208.51' cy='165.70' r='1.87pt' id='svg_f97619c2-9532-4b85-acc2-030cfa5af1d9_el_138' clip-path='url(#svg_f97619c2-9532-4b85-acc2-030cfa5af1d9_cl_3)' fill='#619CFF' fill-opacity='1' stroke='#619CFF' stroke-opacity='1' stroke-width='0.71' stroke-linejoin='round' stroke-linecap='round' title='<b>Species: </b>virginica'/>\n <circle cx='215.86' cy='189.90' r='1.87pt' id='svg_f97619c2-9532-4b85-acc2-030cfa5af1d9_el_139' clip-path='url(#svg_f97619c2-9532-4b85-acc2-030cfa5af1d9_cl_3)' fill='#619CFF' fill-opacity='1' stroke='#619CFF' stroke-opacity='1' stroke-width='0.71' stroke-linejoin='round' stroke-linecap='round' title='<b>Species: </b>virginica'/>\n <circle cx='304.11' cy='93.10' r='1.87pt' id='svg_f97619c2-9532-4b85-acc2-030cfa5af1d9_el_140' clip-path='url(#svg_f97619c2-9532-4b85-acc2-030cfa5af1d9_cl_3)' fill='#619CFF' fill-opacity='1' stroke='#619CFF' stroke-opacity='1' stroke-width='0.71' stroke-linejoin='round' stroke-linecap='round' title='<b>Species: </b>virginica'/>\n <circle cx='304.11' cy='238.30' r='1.87pt' id='svg_f97619c2-9532-4b85-acc2-030cfa5af1d9_el_141' clip-path='url(#svg_f97619c2-9532-4b85-acc2-030cfa5af1d9_cl_3)' fill='#619CFF' fill-opacity='1' stroke='#619CFF' stroke-opacity='1' stroke-width='0.71' stroke-linejoin='round' stroke-linecap='round' title='<b>Species: </b>virginica'/>\n <circle cx='179.09' cy='286.70' r='1.87pt' id='svg_f97619c2-9532-4b85-acc2-030cfa5af1d9_el_142' clip-path='url(#svg_f97619c2-9532-4b85-acc2-030cfa5af1d9_cl_3)' fill='#619CFF' fill-opacity='1' stroke='#619CFF' stroke-opacity='1' stroke-width='0.71' stroke-linejoin='round' stroke-linecap='round' title='<b>Species: </b>virginica'/>\n <circle cx='245.28' cy='165.70' r='1.87pt' id='svg_f97619c2-9532-4b85-acc2-030cfa5af1d9_el_143' clip-path='url(#svg_f97619c2-9532-4b85-acc2-030cfa5af1d9_cl_3)' fill='#619CFF' fill-opacity='1' stroke='#619CFF' stroke-opacity='1' stroke-width='0.71' stroke-linejoin='round' stroke-linecap='round' title='<b>Species: </b>virginica'/>\n <circle cx='149.67' cy='214.10' r='1.87pt' id='svg_f97619c2-9532-4b85-acc2-030cfa5af1d9_el_144' clip-path='url(#svg_f97619c2-9532-4b85-acc2-030cfa5af1d9_cl_3)' fill='#619CFF' fill-opacity='1' stroke='#619CFF' stroke-opacity='1' stroke-width='0.71' stroke-linejoin='round' stroke-linecap='round' title='<b>Species: </b>virginica'/>\n <circle cx='304.11' cy='214.10' r='1.87pt' id='svg_f97619c2-9532-4b85-acc2-030cfa5af1d9_el_145' clip-path='url(#svg_f97619c2-9532-4b85-acc2-030cfa5af1d9_cl_3)' fill='#619CFF' fill-opacity='1' stroke='#619CFF' stroke-opacity='1' stroke-width='0.71' stroke-linejoin='round' stroke-linecap='round' title='<b>Species: </b>virginica'/>\n <circle cx='201.15' cy='226.20' r='1.87pt' id='svg_f97619c2-9532-4b85-acc2-030cfa5af1d9_el_146' clip-path='url(#svg_f97619c2-9532-4b85-acc2-030cfa5af1d9_cl_3)' fill='#619CFF' fill-opacity='1' stroke='#619CFF' stroke-opacity='1' stroke-width='0.71' stroke-linejoin='round' stroke-linecap='round' title='<b>Species: </b>virginica'/>\n <circle cx='230.57' cy='153.60' r='1.87pt' id='svg_f97619c2-9532-4b85-acc2-030cfa5af1d9_el_147' clip-path='url(#svg_f97619c2-9532-4b85-acc2-030cfa5af1d9_cl_3)' fill='#619CFF' fill-opacity='1' stroke='#619CFF' stroke-opacity='1' stroke-width='0.71' stroke-linejoin='round' stroke-linecap='round' title='<b>Species: </b>virginica'/>\n <circle cx='267.34' cy='165.70' r='1.87pt' id='svg_f97619c2-9532-4b85-acc2-030cfa5af1d9_el_148' clip-path='url(#svg_f97619c2-9532-4b85-acc2-030cfa5af1d9_cl_3)' fill='#619CFF' fill-opacity='1' stroke='#619CFF' stroke-opacity='1' stroke-width='0.71' stroke-linejoin='round' stroke-linecap='round' title='<b>Species: </b>virginica'/>\n <circle cx='193.80' cy='214.10' r='1.87pt' id='svg_f97619c2-9532-4b85-acc2-030cfa5af1d9_el_149' clip-path='url(#svg_f97619c2-9532-4b85-acc2-030cfa5af1d9_cl_3)' fill='#619CFF' fill-opacity='1' stroke='#619CFF' stroke-opacity='1' stroke-width='0.71' stroke-linejoin='round' stroke-linecap='round' title='<b>Species: </b>virginica'/>\n <circle cx='186.44' cy='189.90' r='1.87pt' id='svg_f97619c2-9532-4b85-acc2-030cfa5af1d9_el_150' clip-path='url(#svg_f97619c2-9532-4b85-acc2-030cfa5af1d9_cl_3)' fill='#619CFF' fill-opacity='1' stroke='#619CFF' stroke-opacity='1' stroke-width='0.71' stroke-linejoin='round' stroke-linecap='round' title='<b>Species: </b>virginica'/>\n <circle cx='208.51' cy='214.10' r='1.87pt' id='svg_f97619c2-9532-4b85-acc2-030cfa5af1d9_el_151' clip-path='url(#svg_f97619c2-9532-4b85-acc2-030cfa5af1d9_cl_3)' fill='#619CFF' fill-opacity='1' stroke='#619CFF' stroke-opacity='1' stroke-width='0.71' stroke-linejoin='round' stroke-linecap='round' title='<b>Species: </b>virginica'/>\n <circle cx='267.34' cy='189.90' r='1.87pt' id='svg_f97619c2-9532-4b85-acc2-030cfa5af1d9_el_152' clip-path='url(#svg_f97619c2-9532-4b85-acc2-030cfa5af1d9_cl_3)' fill='#619CFF' fill-opacity='1' stroke='#619CFF' stroke-opacity='1' stroke-width='0.71' stroke-linejoin='round' stroke-linecap='round' title='<b>Species: </b>virginica'/>\n <circle cx='282.05' cy='214.10' r='1.87pt' id='svg_f97619c2-9532-4b85-acc2-030cfa5af1d9_el_153' clip-path='url(#svg_f97619c2-9532-4b85-acc2-030cfa5af1d9_cl_3)' fill='#619CFF' fill-opacity='1' stroke='#619CFF' stroke-opacity='1' stroke-width='0.71' stroke-linejoin='round' stroke-linecap='round' title='<b>Species: </b>virginica'/>\n <circle cx='318.82' cy='93.10' r='1.87pt' id='svg_f97619c2-9532-4b85-acc2-030cfa5af1d9_el_154' clip-path='url(#svg_f97619c2-9532-4b85-acc2-030cfa5af1d9_cl_3)' fill='#619CFF' fill-opacity='1' stroke='#619CFF' stroke-opacity='1' stroke-width='0.71' stroke-linejoin='round' stroke-linecap='round' title='<b>Species: </b>virginica'/>\n <circle cx='208.51' cy='214.10' r='1.87pt' id='svg_f97619c2-9532-4b85-acc2-030cfa5af1d9_el_155' clip-path='url(#svg_f97619c2-9532-4b85-acc2-030cfa5af1d9_cl_3)' fill='#619CFF' fill-opacity='1' stroke='#619CFF' stroke-opacity='1' stroke-width='0.71' stroke-linejoin='round' stroke-linecap='round' title='<b>Species: </b>virginica'/>\n <circle cx='201.15' cy='214.10' r='1.87pt' id='svg_f97619c2-9532-4b85-acc2-030cfa5af1d9_el_156' clip-path='url(#svg_f97619c2-9532-4b85-acc2-030cfa5af1d9_cl_3)' fill='#619CFF' fill-opacity='1' stroke='#619CFF' stroke-opacity='1' stroke-width='0.71' stroke-linejoin='round' stroke-linecap='round' title='<b>Species: </b>virginica'/>\n <circle cx='186.44' cy='238.30' r='1.87pt' id='svg_f97619c2-9532-4b85-acc2-030cfa5af1d9_el_157' clip-path='url(#svg_f97619c2-9532-4b85-acc2-030cfa5af1d9_cl_3)' fill='#619CFF' fill-opacity='1' stroke='#619CFF' stroke-opacity='1' stroke-width='0.71' stroke-linejoin='round' stroke-linecap='round' title='<b>Species: </b>virginica'/>\n <circle cx='304.11' cy='189.90' r='1.87pt' id='svg_f97619c2-9532-4b85-acc2-030cfa5af1d9_el_158' clip-path='url(#svg_f97619c2-9532-4b85-acc2-030cfa5af1d9_cl_3)' fill='#619CFF' fill-opacity='1' stroke='#619CFF' stroke-opacity='1' stroke-width='0.71' stroke-linejoin='round' stroke-linecap='round' title='<b>Species: </b>virginica'/>\n <circle cx='201.15' cy='141.50' r='1.87pt' id='svg_f97619c2-9532-4b85-acc2-030cfa5af1d9_el_159' clip-path='url(#svg_f97619c2-9532-4b85-acc2-030cfa5af1d9_cl_3)' fill='#619CFF' fill-opacity='1' stroke='#619CFF' stroke-opacity='1' stroke-width='0.71' stroke-linejoin='round' stroke-linecap='round' title='<b>Species: </b>virginica'/>\n <circle cx='208.51' cy='177.80' r='1.87pt' id='svg_f97619c2-9532-4b85-acc2-030cfa5af1d9_el_160' clip-path='url(#svg_f97619c2-9532-4b85-acc2-030cfa5af1d9_cl_3)' fill='#619CFF' fill-opacity='1' stroke='#619CFF' stroke-opacity='1' stroke-width='0.71' stroke-linejoin='round' stroke-linecap='round' title='<b>Species: </b>virginica'/>\n <circle cx='179.09' cy='189.90' r='1.87pt' id='svg_f97619c2-9532-4b85-acc2-030cfa5af1d9_el_161' clip-path='url(#svg_f97619c2-9532-4b85-acc2-030cfa5af1d9_cl_3)' fill='#619CFF' fill-opacity='1' stroke='#619CFF' stroke-opacity='1' stroke-width='0.71' stroke-linejoin='round' stroke-linecap='round' title='<b>Species: </b>virginica'/>\n <circle cx='245.28' cy='177.80' r='1.87pt' id='svg_f97619c2-9532-4b85-acc2-030cfa5af1d9_el_162' clip-path='url(#svg_f97619c2-9532-4b85-acc2-030cfa5af1d9_cl_3)' fill='#619CFF' fill-opacity='1' stroke='#619CFF' stroke-opacity='1' stroke-width='0.71' stroke-linejoin='round' stroke-linecap='round' title='<b>Species: </b>virginica'/>\n <circle cx='230.57' cy='177.80' r='1.87pt' id='svg_f97619c2-9532-4b85-acc2-030cfa5af1d9_el_163' clip-path='url(#svg_f97619c2-9532-4b85-acc2-030cfa5af1d9_cl_3)' fill='#619CFF' fill-opacity='1' stroke='#619CFF' stroke-opacity='1' stroke-width='0.71' stroke-linejoin='round' stroke-linecap='round' title='<b>Species: </b>virginica'/>\n <circle cx='245.28' cy='177.80' r='1.87pt' id='svg_f97619c2-9532-4b85-acc2-030cfa5af1d9_el_164' clip-path='url(#svg_f97619c2-9532-4b85-acc2-030cfa5af1d9_cl_3)' fill='#619CFF' fill-opacity='1' stroke='#619CFF' stroke-opacity='1' stroke-width='0.71' stroke-linejoin='round' stroke-linecap='round' title='<b>Species: </b>virginica'/>\n <circle cx='164.38' cy='226.20' r='1.87pt' id='svg_f97619c2-9532-4b85-acc2-030cfa5af1d9_el_165' clip-path='url(#svg_f97619c2-9532-4b85-acc2-030cfa5af1d9_cl_3)' fill='#619CFF' fill-opacity='1' stroke='#619CFF' stroke-opacity='1' stroke-width='0.71' stroke-linejoin='round' stroke-linecap='round' title='<b>Species: </b>virginica'/>\n <circle cx='237.92' cy='165.70' r='1.87pt' id='svg_f97619c2-9532-4b85-acc2-030cfa5af1d9_el_166' clip-path='url(#svg_f97619c2-9532-4b85-acc2-030cfa5af1d9_cl_3)' fill='#619CFF' fill-opacity='1' stroke='#619CFF' stroke-opacity='1' stroke-width='0.71' stroke-linejoin='round' stroke-linecap='round' title='<b>Species: </b>virginica'/>\n <circle cx='230.57' cy='153.60' r='1.87pt' id='svg_f97619c2-9532-4b85-acc2-030cfa5af1d9_el_167' clip-path='url(#svg_f97619c2-9532-4b85-acc2-030cfa5af1d9_cl_3)' fill='#619CFF' fill-opacity='1' stroke='#619CFF' stroke-opacity='1' stroke-width='0.71' stroke-linejoin='round' stroke-linecap='round' title='<b>Species: </b>virginica'/>\n <circle cx='230.57' cy='189.90' r='1.87pt' id='svg_f97619c2-9532-4b85-acc2-030cfa5af1d9_el_168' clip-path='url(#svg_f97619c2-9532-4b85-acc2-030cfa5af1d9_cl_3)' fill='#619CFF' fill-opacity='1' stroke='#619CFF' stroke-opacity='1' stroke-width='0.71' stroke-linejoin='round' stroke-linecap='round' title='<b>Species: </b>virginica'/>\n <circle cx='201.15' cy='250.40' r='1.87pt' id='svg_f97619c2-9532-4b85-acc2-030cfa5af1d9_el_169' clip-path='url(#svg_f97619c2-9532-4b85-acc2-030cfa5af1d9_cl_3)' fill='#619CFF' fill-opacity='1' stroke='#619CFF' stroke-opacity='1' stroke-width='0.71' stroke-linejoin='round' stroke-linecap='round' title='<b>Species: </b>virginica'/>\n <circle cx='215.86' cy='189.90' r='1.87pt' id='svg_f97619c2-9532-4b85-acc2-030cfa5af1d9_el_170' clip-path='url(#svg_f97619c2-9532-4b85-acc2-030cfa5af1d9_cl_3)' fill='#619CFF' fill-opacity='1' stroke='#619CFF' stroke-opacity='1' stroke-width='0.71' stroke-linejoin='round' stroke-linecap='round' title='<b>Species: </b>virginica'/>\n <circle cx='193.80' cy='141.50' r='1.87pt' id='svg_f97619c2-9532-4b85-acc2-030cfa5af1d9_el_171' clip-path='url(#svg_f97619c2-9532-4b85-acc2-030cfa5af1d9_cl_3)' fill='#619CFF' fill-opacity='1' stroke='#619CFF' stroke-opacity='1' stroke-width='0.71' stroke-linejoin='round' stroke-linecap='round' title='<b>Species: </b>virginica'/>\n <circle cx='171.74' cy='189.90' r='1.87pt' id='svg_f97619c2-9532-4b85-acc2-030cfa5af1d9_el_172' clip-path='url(#svg_f97619c2-9532-4b85-acc2-030cfa5af1d9_cl_3)' fill='#619CFF' fill-opacity='1' stroke='#619CFF' stroke-opacity='1' stroke-width='0.71' stroke-linejoin='round' stroke-linecap='round' title='<b>Species: </b>virginica'/>\n <rect x='40.84' y='5.98' width='291.22' height='319.44' id='svg_f97619c2-9532-4b85-acc2-030cfa5af1d9_el_173' clip-path='url(#svg_f97619c2-9532-4b85-acc2-030cfa5af1d9_cl_3)' fill='none' stroke='#333333' stroke-opacity='1' stroke-width='1.16' stroke-linejoin='round' stroke-linecap='round'/>\n <defs>\n <clipPath id='svg_f97619c2-9532-4b85-acc2-030cfa5af1d9_cl_4'>\n <rect x='0.00' y='0.00' width='432.00' height='360.00'/>\n <\/clipPath>\n <\/defs>\n <g clip-path='url(#svg_f97619c2-9532-4b85-acc2-030cfa5af1d9_cl_4)'>\n <text x='20.21' y='314.40' id='svg_f97619c2-9532-4b85-acc2-030cfa5af1d9_el_174' font-size='7.20pt' fill='#4D4D4D' fill-opacity='1' font-family='DejaVu Sans'>2.0<\/text>\n <\/g>\n <g clip-path='url(#svg_f97619c2-9532-4b85-acc2-030cfa5af1d9_cl_4)'>\n <text x='20.21' y='253.90' id='svg_f97619c2-9532-4b85-acc2-030cfa5af1d9_el_175' font-size='7.20pt' fill='#4D4D4D' fill-opacity='1' font-family='DejaVu Sans'>2.5<\/text>\n <\/g>\n <g clip-path='url(#svg_f97619c2-9532-4b85-acc2-030cfa5af1d9_cl_4)'>\n <text x='20.21' y='193.40' id='svg_f97619c2-9532-4b85-acc2-030cfa5af1d9_el_176' font-size='7.20pt' fill='#4D4D4D' fill-opacity='1' font-family='DejaVu Sans'>3.0<\/text>\n <\/g>\n <g clip-path='url(#svg_f97619c2-9532-4b85-acc2-030cfa5af1d9_cl_4)'>\n <text x='20.21' y='132.89' id='svg_f97619c2-9532-4b85-acc2-030cfa5af1d9_el_177' font-size='7.20pt' fill='#4D4D4D' fill-opacity='1' font-family='DejaVu Sans'>3.5<\/text>\n <\/g>\n <g clip-path='url(#svg_f97619c2-9532-4b85-acc2-030cfa5af1d9_cl_4)'>\n <text x='20.21' y='72.39' id='svg_f97619c2-9532-4b85-acc2-030cfa5af1d9_el_178' font-size='7.20pt' fill='#4D4D4D' fill-opacity='1' font-family='DejaVu Sans'>4.0<\/text>\n <\/g>\n <g clip-path='url(#svg_f97619c2-9532-4b85-acc2-030cfa5af1d9_cl_4)'>\n <text x='20.21' y='11.89' id='svg_f97619c2-9532-4b85-acc2-030cfa5af1d9_el_179' font-size='7.20pt' fill='#4D4D4D' fill-opacity='1' font-family='DejaVu Sans'>4.5<\/text>\n <\/g>\n <polyline points='37.85,310.90 40.84,310.90' id='svg_f97619c2-9532-4b85-acc2-030cfa5af1d9_el_180' clip-path='url(#svg_f97619c2-9532-4b85-acc2-030cfa5af1d9_cl_4)' fill='none' stroke='#333333' stroke-opacity='1' stroke-width='1.16' stroke-linejoin='round' stroke-linecap='butt'/>\n <polyline points='37.85,250.40 40.84,250.40' id='svg_f97619c2-9532-4b85-acc2-030cfa5af1d9_el_181' clip-path='url(#svg_f97619c2-9532-4b85-acc2-030cfa5af1d9_cl_4)' fill='none' stroke='#333333' stroke-opacity='1' stroke-width='1.16' stroke-linejoin='round' stroke-linecap='butt'/>\n <polyline points='37.85,189.90 40.84,189.90' id='svg_f97619c2-9532-4b85-acc2-030cfa5af1d9_el_182' clip-path='url(#svg_f97619c2-9532-4b85-acc2-030cfa5af1d9_cl_4)' fill='none' stroke='#333333' stroke-opacity='1' stroke-width='1.16' stroke-linejoin='round' stroke-linecap='butt'/>\n <polyline points='37.85,129.40 40.84,129.40' id='svg_f97619c2-9532-4b85-acc2-030cfa5af1d9_el_183' clip-path='url(#svg_f97619c2-9532-4b85-acc2-030cfa5af1d9_cl_4)' fill='none' stroke='#333333' stroke-opacity='1' stroke-width='1.16' stroke-linejoin='round' stroke-linecap='butt'/>\n <polyline points='37.85,68.90 40.84,68.90' id='svg_f97619c2-9532-4b85-acc2-030cfa5af1d9_el_184' clip-path='url(#svg_f97619c2-9532-4b85-acc2-030cfa5af1d9_cl_4)' fill='none' stroke='#333333' stroke-opacity='1' stroke-width='1.16' stroke-linejoin='round' stroke-linecap='butt'/>\n <polyline points='37.85,8.40 40.84,8.40' id='svg_f97619c2-9532-4b85-acc2-030cfa5af1d9_el_185' clip-path='url(#svg_f97619c2-9532-4b85-acc2-030cfa5af1d9_cl_4)' fill='none' stroke='#333333' stroke-opacity='1' stroke-width='1.16' stroke-linejoin='round' stroke-linecap='butt'/>\n <polyline points='105.55,328.41 105.55,325.42' id='svg_f97619c2-9532-4b85-acc2-030cfa5af1d9_el_186' clip-path='url(#svg_f97619c2-9532-4b85-acc2-030cfa5af1d9_cl_4)' fill='none' stroke='#333333' stroke-opacity='1' stroke-width='1.16' stroke-linejoin='round' stroke-linecap='butt'/>\n <polyline points='179.09,328.41 179.09,325.42' id='svg_f97619c2-9532-4b85-acc2-030cfa5af1d9_el_187' clip-path='url(#svg_f97619c2-9532-4b85-acc2-030cfa5af1d9_cl_4)' fill='none' stroke='#333333' stroke-opacity='1' stroke-width='1.16' stroke-linejoin='round' stroke-linecap='butt'/>\n <polyline points='252.63,328.41 252.63,325.42' id='svg_f97619c2-9532-4b85-acc2-030cfa5af1d9_el_188' clip-path='url(#svg_f97619c2-9532-4b85-acc2-030cfa5af1d9_cl_4)' fill='none' stroke='#333333' stroke-opacity='1' stroke-width='1.16' stroke-linejoin='round' stroke-linecap='butt'/>\n <polyline points='326.17,328.41 326.17,325.42' id='svg_f97619c2-9532-4b85-acc2-030cfa5af1d9_el_189' clip-path='url(#svg_f97619c2-9532-4b85-acc2-030cfa5af1d9_cl_4)' fill='none' stroke='#333333' stroke-opacity='1' stroke-width='1.16' stroke-linejoin='round' stroke-linecap='butt'/>\n <g clip-path='url(#svg_f97619c2-9532-4b85-acc2-030cfa5af1d9_cl_4)'>\n <text x='102.50' y='337.79' id='svg_f97619c2-9532-4b85-acc2-030cfa5af1d9_el_190' font-size='7.20pt' fill='#4D4D4D' fill-opacity='1' font-family='DejaVu Sans'>5<\/text>\n <\/g>\n <g clip-path='url(#svg_f97619c2-9532-4b85-acc2-030cfa5af1d9_cl_4)'>\n <text x='176.04' y='337.79' id='svg_f97619c2-9532-4b85-acc2-030cfa5af1d9_el_191' font-size='7.20pt' fill='#4D4D4D' fill-opacity='1' font-family='DejaVu Sans'>6<\/text>\n <\/g>\n <g clip-path='url(#svg_f97619c2-9532-4b85-acc2-030cfa5af1d9_cl_4)'>\n <text x='249.58' y='337.79' id='svg_f97619c2-9532-4b85-acc2-030cfa5af1d9_el_192' font-size='7.20pt' fill='#4D4D4D' fill-opacity='1' font-family='DejaVu Sans'>7<\/text>\n <\/g>\n <g clip-path='url(#svg_f97619c2-9532-4b85-acc2-030cfa5af1d9_cl_4)'>\n <text x='323.12' y='337.79' id='svg_f97619c2-9532-4b85-acc2-030cfa5af1d9_el_193' font-size='7.20pt' fill='#4D4D4D' fill-opacity='1' font-family='DejaVu Sans'>8<\/text>\n <\/g>\n <g clip-path='url(#svg_f97619c2-9532-4b85-acc2-030cfa5af1d9_cl_4)'>\n <text x='147.17' y='351.52' id='svg_f97619c2-9532-4b85-acc2-030cfa5af1d9_el_194' font-size='9.00pt' font-family='DejaVu Sans'>Sepal.Length<\/text>\n <\/g>\n <g clip-path='url(#svg_f97619c2-9532-4b85-acc2-030cfa5af1d9_cl_4)'>\n <text transform='translate(14.73,201.70) rotate(-90.00)' id='svg_f97619c2-9532-4b85-acc2-030cfa5af1d9_el_195' font-size='9.00pt' font-family='DejaVu Sans'>Sepal.Width<\/text>\n <\/g>\n <rect x='344.01' y='125.19' width='82.01' height='81.02' id='svg_f97619c2-9532-4b85-acc2-030cfa5af1d9_el_196' clip-path='url(#svg_f97619c2-9532-4b85-acc2-030cfa5af1d9_cl_4)' fill='#FFFFFF' fill-opacity='1' stroke='none'/>\n <g clip-path='url(#svg_f97619c2-9532-4b85-acc2-030cfa5af1d9_cl_4)'>\n <text x='349.99' y='141.16' id='svg_f97619c2-9532-4b85-acc2-030cfa5af1d9_el_197' font-size='9.00pt' font-family='DejaVu Sans'>Species<\/text>\n <\/g>\n <rect x='349.99' y='148.39' width='17.28' height='17.28' id='svg_f97619c2-9532-4b85-acc2-030cfa5af1d9_el_198' clip-path='url(#svg_f97619c2-9532-4b85-acc2-030cfa5af1d9_cl_4)' fill='#FFFFFF' fill-opacity='1' stroke='none'/>\n <circle cx='358.63' cy='157.03' r='1.87pt' id='svg_f97619c2-9532-4b85-acc2-030cfa5af1d9_el_199' clip-path='url(#svg_f97619c2-9532-4b85-acc2-030cfa5af1d9_cl_4)' fill='#F8766D' fill-opacity='1' stroke='#F8766D' stroke-opacity='1' stroke-width='0.71' stroke-linejoin='round' stroke-linecap='round'/>\n <rect x='349.99' y='165.67' width='17.28' height='17.28' id='svg_f97619c2-9532-4b85-acc2-030cfa5af1d9_el_200' clip-path='url(#svg_f97619c2-9532-4b85-acc2-030cfa5af1d9_cl_4)' fill='#FFFFFF' fill-opacity='1' stroke='none'/>\n <circle cx='358.63' cy='174.31' r='1.87pt' id='svg_f97619c2-9532-4b85-acc2-030cfa5af1d9_el_201' clip-path='url(#svg_f97619c2-9532-4b85-acc2-030cfa5af1d9_cl_4)' fill='#00BA38' fill-opacity='1' stroke='#00BA38' stroke-opacity='1' stroke-width='0.71' stroke-linejoin='round' stroke-linecap='round'/>\n <rect x='349.99' y='182.95' width='17.28' height='17.28' id='svg_f97619c2-9532-4b85-acc2-030cfa5af1d9_el_202' clip-path='url(#svg_f97619c2-9532-4b85-acc2-030cfa5af1d9_cl_4)' fill='#FFFFFF' fill-opacity='1' stroke='none'/>\n <circle cx='358.63' cy='191.59' r='1.87pt' id='svg_f97619c2-9532-4b85-acc2-030cfa5af1d9_el_203' clip-path='url(#svg_f97619c2-9532-4b85-acc2-030cfa5af1d9_cl_4)' fill='#619CFF' fill-opacity='1' stroke='#619CFF' stroke-opacity='1' stroke-width='0.71' stroke-linejoin='round' stroke-linecap='round'/>\n <g clip-path='url(#svg_f97619c2-9532-4b85-acc2-030cfa5af1d9_cl_4)'>\n <text x='373.24' y='160.53' id='svg_f97619c2-9532-4b85-acc2-030cfa5af1d9_el_204' font-size='7.20pt' font-family='DejaVu Sans'>setosa<\/text>\n <\/g>\n <g clip-path='url(#svg_f97619c2-9532-4b85-acc2-030cfa5af1d9_cl_4)'>\n <text x='373.24' y='177.81' id='svg_f97619c2-9532-4b85-acc2-030cfa5af1d9_el_205' font-size='7.20pt' font-family='DejaVu Sans'>versicolor<\/text>\n <\/g>\n <g clip-path='url(#svg_f97619c2-9532-4b85-acc2-030cfa5af1d9_cl_4)'>\n <text x='373.24' y='195.09' id='svg_f97619c2-9532-4b85-acc2-030cfa5af1d9_el_206' font-size='7.20pt' font-family='DejaVu Sans'>virginica<\/text>\n <\/g>\n <\/g>\n<\/svg>","js":null,"uid":"svg_f97619c2-9532-4b85-acc2-030cfa5af1d9","ratio":1.2,"settings":{"tooltip":{"css":".tooltip_SVGID_ { padding:5px;background:black;color:white;border-radius:2px 2px 2px 2px;text-align:left; ; position:absolute;pointer-events:none;z-index:999;}\n","placement":"container","offx":10,"offy":0,"use_cursor_pos":true,"opacity":0.9,"usefill":false,"usestroke":false,"delay":{"over":200,"out":500}},"hover":{"css":".hover_SVGID_ { fill:orange;stroke:gray; }","reactive":false},"hoverkey":{"css":".hover_key_SVGID_ { stroke:red; }","reactive":false},"hovertheme":{"css":".hover_theme_SVGID_ { fill:green; }","reactive":false},"hoverinv":{"css":""},"zoom":{"min":1,"max":1},"capture":{"css":".selected_SVGID_ { fill:red;stroke:gray; }","type":"multiple","only_shiny":true,"selected":[]},"capturekey":{"css":".selected_key_SVGID_ { stroke:gray; }","type":"single","only_shiny":true,"selected":[]},"capturetheme":{"css":".selected_theme_SVGID_ { stroke:gray; }","type":"single","only_shiny":true,"selected":[]},"toolbar":{"position":"top","saveaspng":false,"pngname":"diagram"},"sizing":{"rescale":true,"width":0.75}}},"evals":[],"jsHooks":[]}</script> ] ??? Most interactive plotting libraries are not as complete as `ggplot2`. Therefore, some packages explore ways of converting `ggplot2` objects into interactive graphics --- name: extension class: spaced ## Extensions * [**gridExtra**](https://cran.r-project.org/web/packages/gridExtra/index.html): Extends grid graphics functionality * [**ggpubr**](http://www.sthda.com/english/rpkgs/ggpubr/): Useful functions to prepare plots for publication * [**cowplot**](https://cran.r-project.org/web/packages/cowplot/vignettes/introduction.html): Combining plots * [**ggthemes**](https://cran.r-project.org/web/packages/ggthemes/vignettes/ggthemes.html): Set of extra themes * [**ggthemr**](https://github.com/cttobin/ggthemr): More themes * [**ggsci**](https://cran.r-project.org/web/packages/ggsci/vignettes/ggsci.html): Color palettes for scales * [**ggrepel**](https://cran.r-project.org/web/packages/ggrepel/vignettes/ggrepel.html): Advanced text labels including overlap control * [**ggmap**](https://github.com/dkahle/ggmap): Dedicated to mapping * [**ggraph**](https://github.com/thomasp85/ggraph): Network graphs * [**ggiraph**](http://davidgohel.github.io/ggiraph/): Converting ggplot2 to interactive graphics A collection of ggplot extension packages: [https://exts.ggplot2.tidyverse.org/](https://exts.ggplot2.tidyverse.org/). --- name: help class: spaced ## Help * [**ggplot2 official reference**](http://ggplot2.tidyverse.org/reference/) * [**The R cookbook**](http://www.cookbook-r.com/) * [**StackOverflow**](https://stackoverflow.com/) * [**RStudio Cheatsheet**](https://www.rstudio.com/resources/cheatsheets/) * [**r-statistics Cheatsheet**](http://r-statistics.co/ggplot2-cheatsheet.html) * [**ggplot2 GUI**](https://site.shinyserver.dck.gmw.rug.nl/ggplotgui/) * Numerous personal blogs, r-bloggers.com etc. <!-- --------------------- Do not edit this and below --------------------- --> --- name: end-slide class: end-slide, middle count: false # Thank you. Questions? <p>R version 4.0.2 (2020-06-22)<br><p>Platform: x86_64-conda_cos6-linux-gnu (64-bit)</p><p>OS: Ubuntu 20.04.2 LTS</p><br> Built on : <i class='fa fa-calendar' aria-hidden='true'></i> 14-Jun-2021 at <i class='fa fa-clock-o' aria-hidden='true'></i> 12:58:36 __2019__ • [SciLifeLab](https://www.scilifelab.se/) • [NBIS](https://nbis.se/)