class: center, middle, inverse, title-slide # Mathematical Statistics in R ## Advanced R for Bioinformatics. Visby, 2018. ### Nikolay Oskolkov ### 15 Juni, 2018 --- class: spaced ## What is Mathematical Statistics? * Can Mathematical Statistics mean * a statistical test? * a probability distribution? * or maybe a p-value? * Classic statistics **is not the only way** to analyze your data  --- name: Statistics ## Different Types of Data Analysis * Depends on the amount of data we have * Balance between the numbers of features and observations 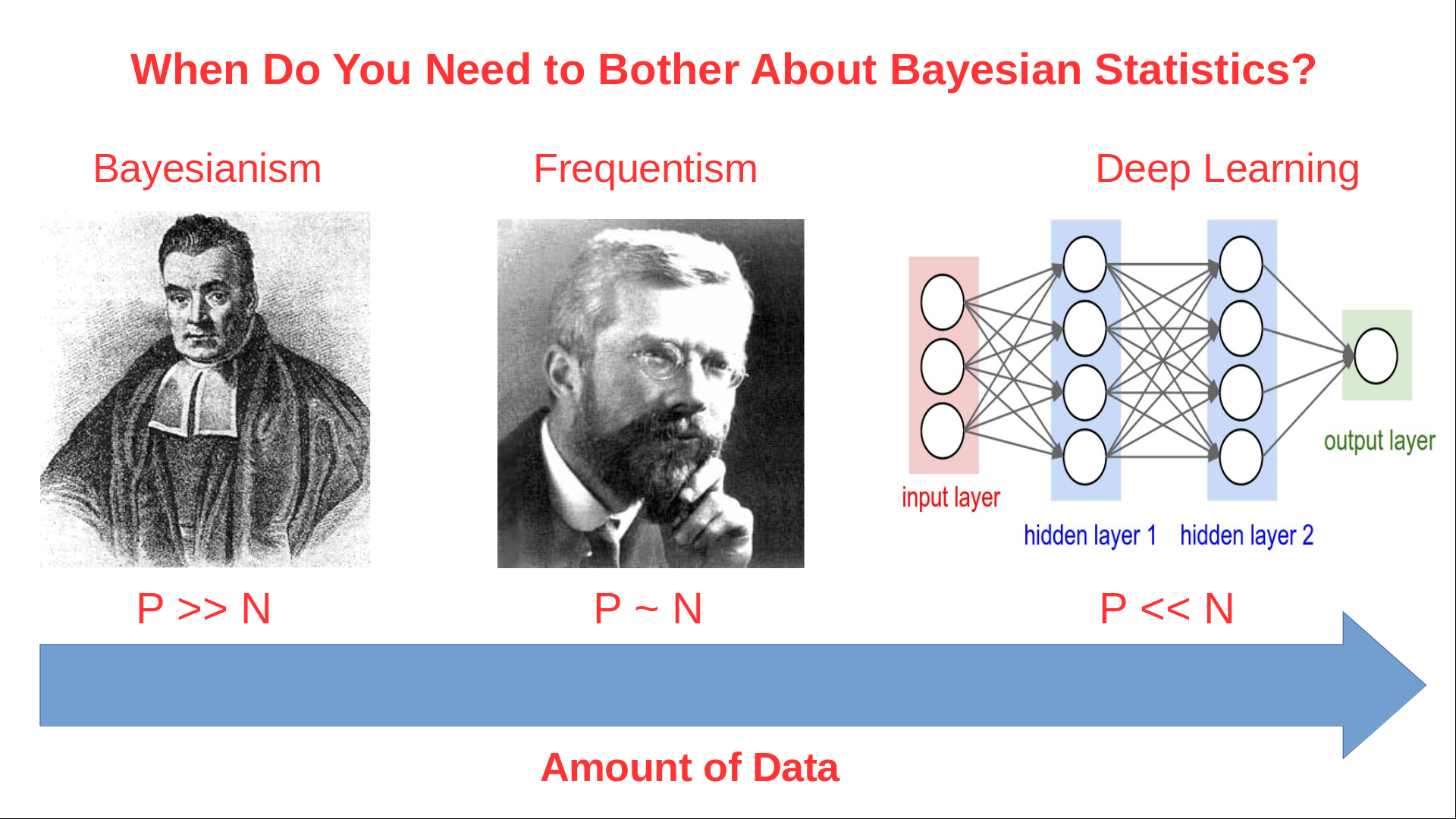 --- name: Types of Analysis ## Frequentist Statistics Failure  --- name: Frequentist Statistics Brain Damaging ## Frequentist Statistics Brain Damaging 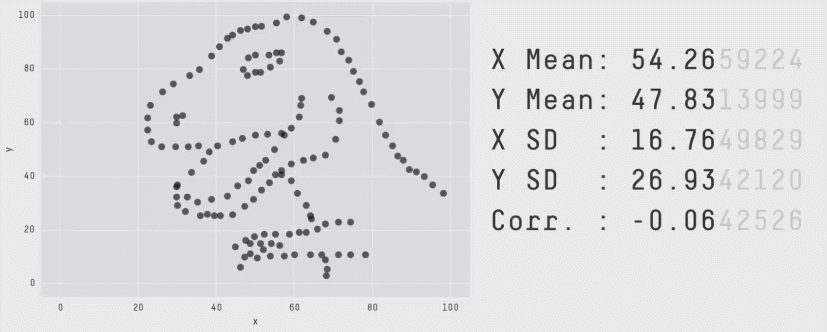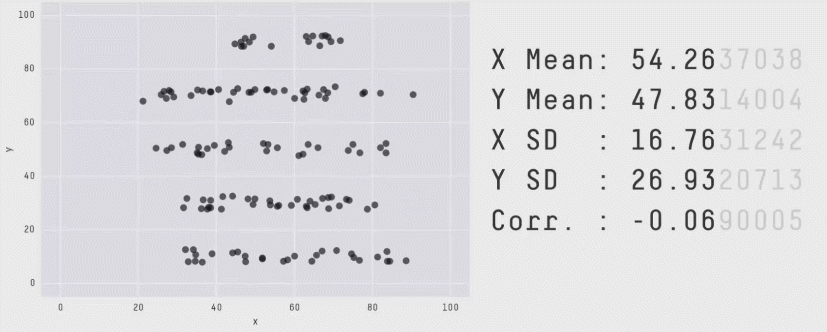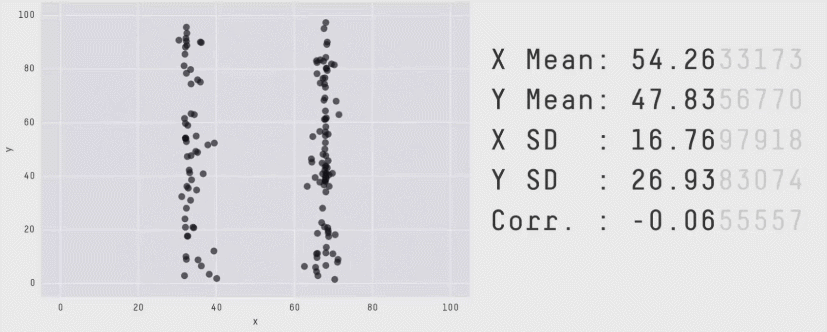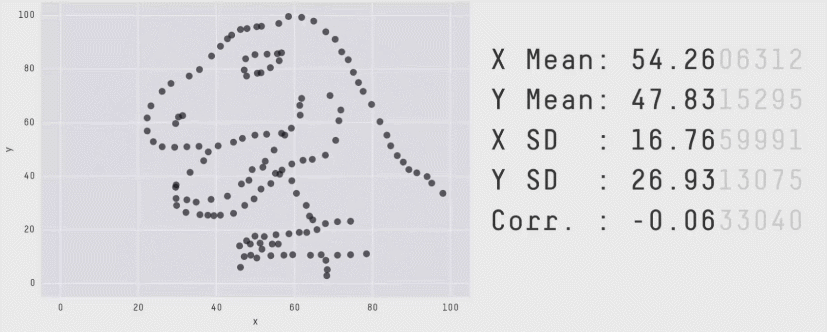  --- name: Frequentist Statistics Brain Damaging (Cont.) ## Maximum Likelihood Principle * We maximize probability to observe the data `\(X_i\)` `$$\rm{L}\,(\,\rm{X_i} \,|\, \mu,\sigma\,) = \prod_{i=0}^{N}\frac{1}{\sqrt{2\pi\sigma²}} \exp^{\displaystyle -\frac{(X_i-\mu)^2}{2\sigma²}}\\ \mu = \frac{1}{N}\sum_{i=0}^N \rm{X_i}\\ \sigma^2 = \frac{1}{N}\sum_{i=0}^N (\rm{X_i}-\mu)^2$$` -- * Maximum Likelihood has many assumptions: * Large sample size * Gaussian distribution * Homoscedasticity * Uncorrelated errors * Convergence of covariance * Those assumptions are not fulfilled in the real world --- name: Maximum Likelihood Principle ## Two-Groups Statistical Test ```r set.seed(12) X<-c(rnorm(20,mean=5,sd=2),12,15,14,16) Y<-c(rnorm(24,mean=7,sd=2)) boxplot(X,Y,ylab="DIFFERENCE",names=c("X","Y")) ``` <img src="Presentation_GeneralStats_files/figure-html/unnamed-chunk-3-1.png" style="display: block; margin: auto auto auto 0;" /> --- name: Statistical Test ## Parametric Statistical Test Fails ```r t.test(X,Y) ``` ``` ## ## Welch Two Sample t-test ## ## data: X and Y ## t = -1.084, df = 31.678, p-value = 0.2865 ## alternative hypothesis: true difference in means is not equal to 0 ## 95 percent confidence interval: ## -2.8819545 0.8804344 ## sample estimates: ## mean of x mean of y ## 5.989652 6.990412 ``` -- .center[ <img src="Presentation_GeneralStats_files/figure-html/unnamed-chunk-5-1.png" style="display: block; margin: auto auto auto 0;" /> ] --- name: Parametric Statistical Test ## Resampling ```r observed <- median(Y)-median(X) print(paste0("Observed difference = ",observed)) res <- vector(length=1000) for(i in 1:1000){Z<-c(X,Y); Y_res<-sample(Z,length(Y),FALSE); X_res<-sample(Z,length(X),FALSE); res[i]<-median(Y_res)-median(X_res)} hist(abs(res), breaks=100, main="Resampled", xlab="Difference") print(paste0("p-value = ", sum(abs(res) >= abs(observed))/1000)) ``` ``` ## [1] "Observed difference = 2.33059085402647" ## [1] "p-value = 0.002" ``` <img src="Presentation_GeneralStats_files/figure-html/unnamed-chunk-6-1.png" style="display: block; margin: auto auto auto 0;" /> --- name: Resampling ## ML Does Not Stand Non-Independence ``` ## n1 n2 n3 n4 n5 ## p1 1.0862296 0.2715666 -1.5000279 -2.1023121 0.47450044 ## p2 0.5871922 1.1193045 -2.4755893 -0.7310311 -0.06784677 ## p3 1.8750843 1.2789147 0.4295881 0.8118376 -0.22511970 ## p4 0.4396084 -0.1569604 -0.3682398 0.8253523 0.23070668 ## p5 -0.4962881 -1.0323406 -0.9390560 -0.4715313 0.35579237 ``` * Two types of non-independence in data * between samples * between features .pull-left-50[ ### Random Effects <img src="Random_Effects.jpg" style="width: 60%;" />] .pull-right-50[ ### Lasso <img src="lasso.jpg" style="width: 40%;" />] --- name: ML Does Not Stand Non-Independence ## Linear Model with Non-Independence ```r library("lme4") library("ggplot2") ggplot(sleepstudy,aes(x=Days,y=Reaction)) + geom_point() + geom_smooth(method="lm") ``` <img src="Presentation_GeneralStats_files/figure-html/unnamed-chunk-8-1.png" style="display: block; margin: auto auto auto 0;" /> --- name: Linear Model with Non-Independence ## Fit Linear Model for Each Individual ```r ggplot(sleepstudy, aes(x = Days, y = Reaction)) + geom_smooth(method = "lm", level = 0.95) + geom_point() + facet_wrap( ~ Subject, nrow = 3, ncol = 6) ``` <img src="Presentation_GeneralStats_files/figure-html/unnamed-chunk-9-1.png" style="display: block; margin: auto auto auto 0;" /> --- name: Fit Linear Model for Each Individual ## Random Effects and Missing Heritability <img src="MissingHeritability.png" style="width: 90%;" /> --- name: The Case of the Missing Heritability ## Random Effects Modelling * Allow individual level Slopes and Intercepts * This is nothing else than Bayesian Priors on coefficients `$$\rm{Reaction} = \alpha_i + \beta_i \rm{Days}$$` ```r lmerfit <- lmer(Reaction ~ Days + (Days | Subject), sleepstudy) ``` .pull-left-50[ <img src="Presentation_GeneralStats_files/figure-html/unnamed-chunk-11-1.png" style="display: block; margin: auto auto auto 0;" /> ] .pull-right-50[ <img src="Presentation_GeneralStats_files/figure-html/unnamed-chunk-12-1.png" style="display: block; margin: auto auto auto 0;" /> ] --- name: Random Effects Modelling ## Linear Mixed Models (LMM) ```r summary(lmer(Reaction ~ Days + (Days | Subject), sleepstudy)) ``` ``` ## Linear mixed model fit by REML ['lmerMod'] ## Formula: Reaction ~ Days + (Days | Subject) ## Data: sleepstudy ## ## REML criterion at convergence: 1743.6 ## ## Scaled residuals: ## Min 1Q Median 3Q Max ## -3.9536 -0.4634 0.0231 0.4634 5.1793 ## ## Random effects: ## Groups Name Variance Std.Dev. Corr ## Subject (Intercept) 612.09 24.740 ## Days 35.07 5.922 0.07 ## Residual 654.94 25.592 ## Number of obs: 180, groups: Subject, 18 ## ## Fixed effects: ## Estimate Std. Error t value ## (Intercept) 251.405 6.825 36.838 ## Days 10.467 1.546 6.771 ## ## Correlation of Fixed Effects: ## (Intr) ## Days -0.138 ``` --- name: Linear Mixed Models (LMM) ## LMM Average Fit <img src="Presentation_GeneralStats_files/figure-html/unnamed-chunk-14-1.png" style="display: block; margin: auto auto auto 0;" /> --- name: LMM Average Fit ## LMM Individual Fit <img src="Presentation_GeneralStats_files/figure-html/unnamed-chunk-15-1.png" style="display: block; margin: auto auto auto 0;" /> --- name: LMM Individual Fit ## What is Bayesian Statistics for You?  .small[ * **Handling Missing Data** * **Handling Non-Gaussian Data** * **Cause or Consequence** * **Lack of Statistical Power** * **Overfitting and Correction for Multiple Testing (FDR)** * **Testing for Significance and P-Value** ] --- name: What is Bayesian Statistics for You? ## Frequentist vs. Bayesian Linear Model .pull-left-50[ ### Maximum Likelihood `$$y = \alpha+\beta x$$` `$$L(y) \sim e^{-\frac{(y-\alpha-\beta x)^2}{2\sigma^2}}$$` `$$\max_{\alpha,\beta,\sigma}L(y) \Longrightarrow \hat\alpha, \hat\beta, \hat\sigma$$` ] -- .pull-right-50[ ### Bayesian Linear Fitting `$$y \sim \it N(\mu,\sigma) \quad\rightarrow \textrm{Likelihood}:L(y| \mu,\sigma)$$` `$$\mu = \alpha + \beta x \quad \textrm{(deterministic)}$$` `$$\alpha \sim \it N(\mu_\alpha,\sigma_\alpha) \quad\rightarrow\textrm{Prior on} \quad\alpha:P(\alpha|\mu_\alpha,\sigma_\alpha)$$` `$$\beta \sim \it N(\mu_\beta,\sigma_\beta) \quad\rightarrow\textrm{Prior on} \quad\beta: P(\beta|\mu_\beta,\sigma_\beta)$$` `$$P(\alpha, \beta|y, \mu_\alpha,\sigma_\alpha,\mu_\beta,\sigma_\beta, \sigma) = \frac{L(y|\mu,\sigma) P(\alpha|\mu_\alpha,\sigma_\alpha)P(\beta|\mu_\beta,\sigma_\beta)}{\int_{\alpha,\beta,\sigma}L(y|\mu,\sigma) P(\alpha|\mu_\alpha,\sigma_\alpha)P(\beta|\mu_\beta,\sigma_\beta)}$$` `$$\max_{\alpha,\beta,\sigma}P(\alpha, \beta, \sigma|y, \mu_\alpha,\sigma_\alpha,\mu_\beta,\sigma_\beta) \Longrightarrow \hat\alpha,\hat\beta,\hat\sigma$$` <!-- `$$y \sim \it N(\mu,\sigma) \quad\textrm{- Likelihood L(y)}$$` --> <!-- `$$\mu = \alpha + \beta x$$` --> <!-- $$\alpha \sim \it N(\mu_\alpha,\sigma_\alpha) \quad\textrm{- Prior on} \quad\alpha \\ --> <!-- \beta \sim \it N(\mu_\beta,\sigma_\beta) \quad\textrm{- Prior on} \quad\beta$$ --> <!-- `$$P(\mu_\alpha,\sigma_\alpha,\mu_\beta,\sigma_\beta,\sigma) \sim L(y)N(\mu_\alpha,\sigma_\alpha)N(\mu_\beta,\sigma_\beta)$$` --> <!-- `$$\max_{\mu_\alpha,\sigma_\alpha,\mu_\beta,\sigma_\beta,\sigma}P(\mu_\alpha,\sigma_\alpha,\mu_\beta,\sigma_\beta,\sigma) \Longrightarrow \hat\mu_\alpha,\hat\sigma_\alpha,\hat\mu_\beta,\hat\sigma_\beta,\hat\sigma$$` --> ] --- name: Frequentist vs. Bayesian Fitting ## Bayesian Linear Model ```r library("brms") options(mc.cores = parallel::detectCores()) brmfit <- brm(Reaction ~ Days + (Days | Subject), data = sleepstudy) summary(brmfit) ``` ``` ## Family: gaussian ## Links: mu = identity; sigma = identity ## Formula: Reaction ~ Days + (Days | Subject) ## Data: sleepstudy (Number of observations: 180) ## Samples: 4 chains, each with iter = 2000; warmup = 1000; thin = 1; ## total post-warmup samples = 4000 ## ICs: LOO = NA; WAIC = NA; R2 = NA ## ## Group-Level Effects: ## ~Subject (Number of levels: 18) ## Estimate Est.Error l-95% CI u-95% CI Eff.Sample Rhat ## sd(Intercept) 26.99 6.84 15.68 42.52 1869 1.00 ## sd(Days) 6.61 1.53 4.19 10.12 1591 1.00 ## cor(Intercept,Days) 0.09 0.30 -0.48 0.68 978 1.00 ## ## Population-Level Effects: ## Estimate Est.Error l-95% CI u-95% CI Eff.Sample Rhat ## Intercept 251.22 7.23 236.75 265.84 1592 1.00 ## Days 10.47 1.72 7.13 13.89 1332 1.00 ## ## Family Specific Parameters: ## Estimate Est.Error l-95% CI u-95% CI Eff.Sample Rhat ## sigma 25.85 1.55 23.08 29.17 4000 1.00 ## ## Samples were drawn using sampling(NUTS). For each parameter, Eff.Sample ## is a crude measure of effective sample size, and Rhat is the potential ## scale reduction factor on split chains (at convergence, Rhat = 1). ``` --- name: Bayesian Linear Model ## Bayesian Population-Level Fit <img src="Presentation_GeneralStats_files/figure-html/unnamed-chunk-17-1.png" style="display: block; margin: auto auto auto 0;" /> --- name: Bayesian Population-Level Fit ## Bayesian Group-Level Fit <img src="Presentation_GeneralStats_files/figure-html/unnamed-chunk-18-1.png" style="display: block; margin: auto auto auto 0;" /> --- name: Bayesian Group-Level Fit ## Feature Non-Independence: LASSO `$$Y = \beta_1X_1+\beta_2X_2+\epsilon \\ \textrm{OLS} = (y-\beta_1X_1-\beta_2X_2)^2 \\ \textrm{Penalized OLS} = (y-\beta_1X_1-\beta_2X_2)^2 + \lambda(|\beta_1|+|\beta_2|)$$` .center[ <img src="CV_lambda.png" style="width: 60%;" />] --- name: Feature Non-Independence: LASSO ## Dimensionality Reduction * Can Dimensionality Reduction mean * PCA? * tSNE? .pull-left-50[ .pull-right-50[ <img src="PCA.png" style="width: 100%;" />]] .pull-right-50[ <img src="tSNE.png" style="width: 50%;" />] -- .pull-left-50[ .pull-right-50[ <img src="DarkMagic.jpg" style="width: 100%;" />]] .pull-right-50[ No, this is about overcoming **The Curse of Dimenionality** also known as Rao's Paradox ] --- name: Dimensionality Reduction ## Low Dimensional Space ```r set.seed(123); n <- 20; p <- 2 Y <- rnorm(n); X <- matrix(rnorm(n*p),n,p); summary(lm(Y~X)) ``` ``` ## ## Call: ## lm(formula = Y ~ X) ## ## Residuals: ## Min 1Q Median 3Q Max ## -2.0522 -0.6380 0.1451 0.3911 1.8829 ## ## Coefficients: ## Estimate Std. Error t value Pr(>|t|) ## (Intercept) 0.14950 0.22949 0.651 0.523 ## X1 -0.09405 0.28245 -0.333 0.743 ## X2 -0.11919 0.24486 -0.487 0.633 ## ## Residual standard error: 1.017 on 17 degrees of freedom ## Multiple R-squared: 0.02204, Adjusted R-squared: -0.09301 ## F-statistic: 0.1916 on 2 and 17 DF, p-value: 0.8274 ``` --- name: Low Dimensional Space ## Going to Higher Dimensions ```r set.seed(123456); n <- 20; p <- 10 Y <- rnorm(n); X <- matrix(rnorm(n*p),n,p); summary(lm(Y~X)) ``` ``` ## ## Call: ## lm(formula = Y ~ X) ## ## Residuals: ## Min 1Q Median 3Q Max ## -1.0255 -0.4320 0.1056 0.4493 1.0617 ## ## Coefficients: ## Estimate Std. Error t value Pr(>|t|) ## (Intercept) 0.54916 0.26472 2.075 0.0679 . ## X1 0.30013 0.21690 1.384 0.1998 ## X2 0.68053 0.27693 2.457 0.0363 * ## X3 -0.10675 0.26010 -0.410 0.6911 ## X4 -0.21367 0.33690 -0.634 0.5417 ## X5 -0.19123 0.31881 -0.600 0.5634 ## X6 0.81074 0.25221 3.214 0.0106 * ## X7 0.09634 0.24143 0.399 0.6992 ## X8 -0.29864 0.19004 -1.571 0.1505 ## X9 -0.78175 0.35408 -2.208 0.0546 . ## X10 0.83736 0.36936 2.267 0.0496 * ## --- ## Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1 ## ## Residual standard error: 0.8692 on 9 degrees of freedom ## Multiple R-squared: 0.6592, Adjusted R-squared: 0.2805 ## F-statistic: 1.741 on 10 and 9 DF, p-value: 0.2089 ``` --- name: Going to Higher Dimensions ## Even Higher Dimensions ```r set.seed(123456); n <- 20; p <- 20 Y <- rnorm(n); X <- matrix(rnorm(n*p),n,p); summary(lm(Y~X)) ``` ``` ## ## Call: ## lm(formula = Y ~ X) ## ## Residuals: ## ALL 20 residuals are 0: no residual degrees of freedom! ## ## Coefficients: (1 not defined because of singularities) ## Estimate Std. Error t value Pr(>|t|) ## (Intercept) 1.34889 NA NA NA ## X1 0.66218 NA NA NA ## X2 0.76212 NA NA NA ## X3 -1.35033 NA NA NA ## X4 -0.57487 NA NA NA ## X5 0.02142 NA NA NA ## X6 0.40290 NA NA NA ## X7 0.03313 NA NA NA ## X8 -0.31983 NA NA NA ## X9 -0.92833 NA NA NA ## X10 0.18091 NA NA NA ## X11 -1.37618 NA NA NA ## X12 2.11438 NA NA NA ## X13 -1.75103 NA NA NA ## X14 -1.55073 NA NA NA ## X15 0.01112 NA NA NA ## X16 -0.50943 NA NA NA ## X17 -0.47576 NA NA NA ## X18 0.31793 NA NA NA ## X19 1.43615 NA NA NA ## X20 NA NA NA NA ## ## Residual standard error: NaN on 0 degrees of freedom ## Multiple R-squared: 1, Adjusted R-squared: NaN ## F-statistic: NaN on 19 and 0 DF, p-value: NA ``` --- name: Even Higher Dimensions ## The Math Blows Up in High Dimensions Let us now take a closer look at why exactly the ML math blows up when n<=p. Consider a linear model: `$$Y = \beta X$$` Let us make a few mathematical tricks in order to get a solution for the coefficients of the linear model: `$$X^TY = \beta X^TX \\ (X^TX)^{-1}X^TY = \beta(X^TX)^{-1} X^TX \\ (X^TX)^{-1}X^TY = \beta$$` The inverse matrix `\((X^TX)^{-1}\)` **diverges** when p=>n as variables in X become correlated --- name: Maximum Likelihood Blows Up ## No True Effects in High Dimensions .pull-left-50[ ```r set.seed(12345) N <- 100 x <- rnorm(N) y <- 2*x+rnorm(N) df <- data.frame(x,y) ``` ] .pull-right-50[ ```r for(i in 1:10) { df[,paste0("PC",i)]<- Covar*(1-i/10)*y+rnorm(N) } ``` ] .center[ <img src="Presentation_GeneralStats_files/figure-html/unnamed-chunk-24-1.png" style="display: block; margin: auto auto auto 0;" /> ] --- name: No True Effects in High Dimensions ## Principal Component Analysis (PCA) * Collapse p features (p>>n) to few latent features and keep variation * Rotation and shift of coordinate system toward maximal variance * PCA is an **eigen matrix decomposition** problem .pull-left-50[ <img src="pca_demo.gif" style="width: 100%;" />] .pull-right-50[ `$$PC = u^T X = X^Tu$$` X is mean centered `\(\Longrightarrow <PC> = 0\)` `$$<(PC-<PC>)^2> = <PC^2> = u^T X X^Tu \\ X X^T=A \\ < PC^2 > = u^T Au$$` A is **variance-covariance** of X `$$\rm{max}(u^T Au + \lambda(1-u^Tu))=0 \\ Au = \lambda u$$` ] --- name: PCA ## PCA on MNIST <img src="Presentation_GeneralStats_files/figure-html/PCA-1.png" style="display: block; margin: auto auto auto 0;" /> --- name: PCA on MNIST ## t-distributed Stochastic Neighbor Embedding (tSNE) .center[ <img src="tSNE_Scheme.png" style="width: 65%;" />] --- name: tSNE ## tSNE on MNIST <img src="Presentation_GeneralStats_files/figure-html/tSNE-1.png" style="display: block; margin: auto auto auto 0;" /> --- name: tSNE on MNIST --- name: end-slide class: end-slide # Thank you